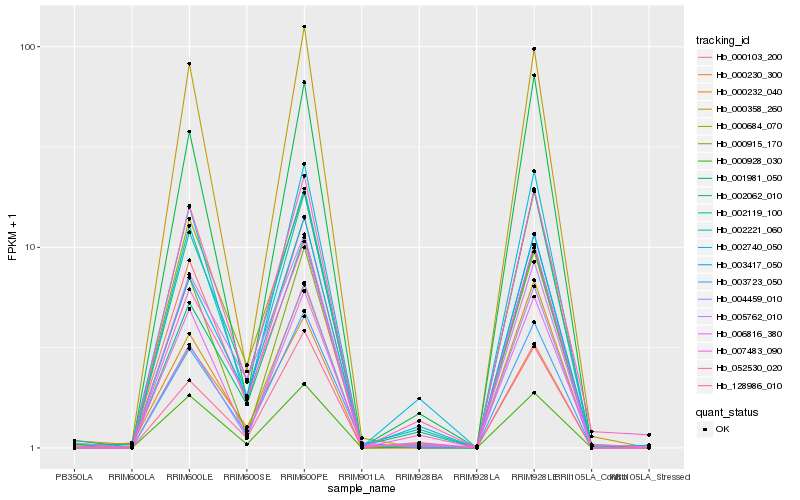
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003723_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002119_100 |
0.0751694076 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 3 |
Hb_000103_200 |
0.0803983037 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_000684_070 |
0.0851072751 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000358_260 |
0.0867246808 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Vitis vinifera] |
| 6 |
Hb_000915_170 |
0.0884651054 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 7 |
Hb_004459_010 |
0.091212 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 8 |
Hb_007483_090 |
0.0915343462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000230_300 |
0.0922149026 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 10 |
Hb_003417_050 |
0.09379886 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_006816_380 |
0.0946034979 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 12 |
Hb_000928_030 |
0.0955863981 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 13 |
Hb_000232_040 |
0.0972673645 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 14 |
Hb_002221_060 |
0.0988605637 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_002740_050 |
0.100798005 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 16 |
Hb_001981_050 |
0.1023490807 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 17 |
Hb_128986_010 |
0.1025725093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002062_010 |
0.1032763303 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 19 |
Hb_005762_010 |
0.1041529855 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 20 |
Hb_052530_020 |
0.1058352711 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |