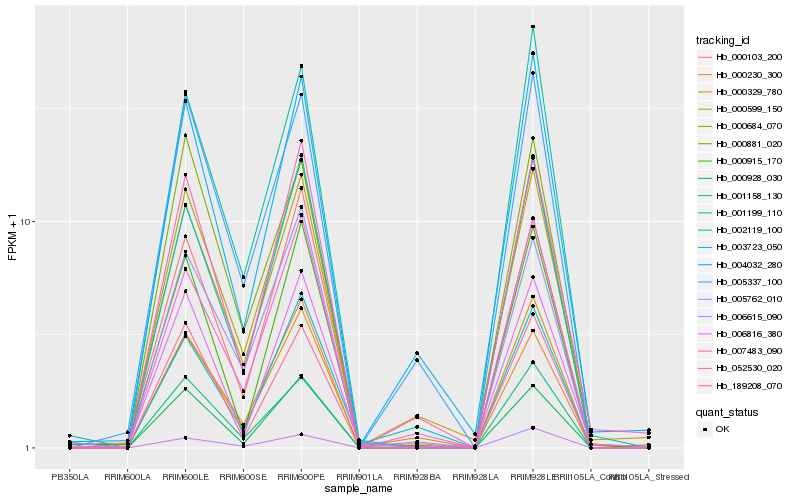
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001158_130 |
0.0618973562 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 3 |
Hb_002119_100 |
0.0749888298 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 4 |
Hb_001199_110 |
0.0787560551 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007483_090 |
0.0791050181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000103_200 |
0.0842042243 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_000599_150 |
0.084371553 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 8 |
Hb_006615_090 |
0.0844384023 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 9 |
Hb_003723_050 |
0.0851072751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_189208_070 |
0.0854917263 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000230_300 |
0.0862980586 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 12 |
Hb_000928_030 |
0.0867014753 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 13 |
Hb_052530_020 |
0.0880550836 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 14 |
Hb_000329_780 |
0.0933934777 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 15 |
Hb_005762_010 |
0.0947737918 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_005337_100 |
0.0974275684 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 17 |
Hb_006816_380 |
0.0986117035 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 18 |
Hb_000881_020 |
0.0995170734 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 19 |
Hb_000915_170 |
0.0998037325 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 20 |
Hb_004032_280 |
0.1028380112 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |