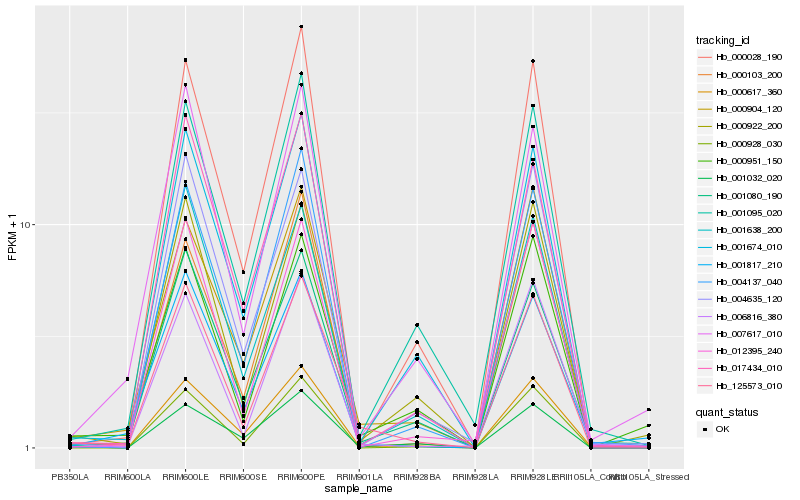
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_125573_010 |
0.0 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_000904_120 |
0.1078872638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012395_240 |
0.1112491461 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000922_200 |
0.1115197376 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 5 |
Hb_001638_200 |
0.1117472215 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 6 |
Hb_006816_380 |
0.1124449014 |
- |
- |
PREDICTED: kinesin-4-like [Jatropha curcas] |
| 7 |
Hb_001080_190 |
0.1142170543 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000617_360 |
0.1153704966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000928_030 |
0.1154146721 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 10 |
Hb_000028_190 |
0.1173587329 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 11 |
Hb_001674_010 |
0.1173693937 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 12 |
Hb_000951_150 |
0.1217103999 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 13 |
Hb_017434_010 |
0.1229744639 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_004635_120 |
0.1229948574 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 15 |
Hb_004137_040 |
0.1233445177 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 16 |
Hb_001032_020 |
0.1242859473 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 17 |
Hb_001817_210 |
0.1244116885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007617_010 |
0.1245814333 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000103_200 |
0.1246807945 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 20 |
Hb_001095_020 |
0.1256691144 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |