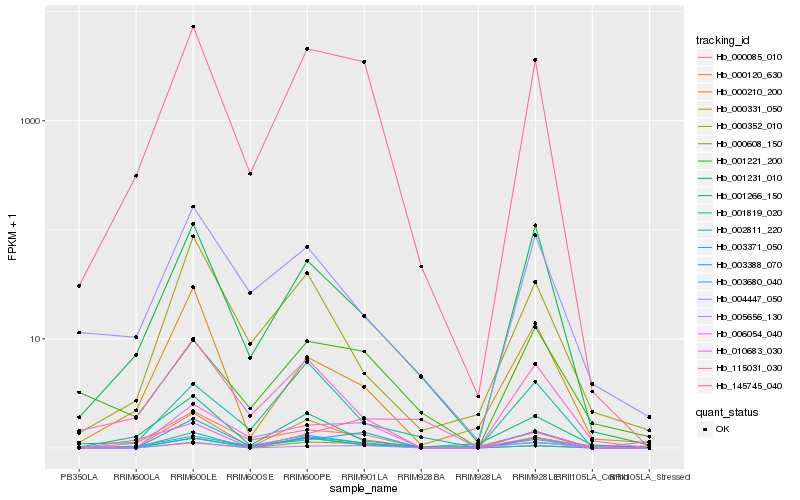
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_145745_040 |
0.0 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 2 |
Hb_000608_150 |
0.177054522 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003371_050 |
0.1817480184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000331_050 |
0.1818045662 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 5 |
Hb_000120_630 |
0.1830060478 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 6 |
Hb_003388_070 |
0.1871473614 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 7 |
Hb_000085_010 |
0.2181378583 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_001231_010 |
0.233379031 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 9 |
Hb_115031_030 |
0.2435960071 |
- |
- |
ribosomal protein S12 (chloroplast) [Camellia sinensis] |
| 10 |
Hb_003680_040 |
0.2449211763 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005656_130 |
0.2518006459 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 12 |
Hb_002811_220 |
0.2528316948 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_006054_040 |
0.2553293475 |
- |
- |
- |
| 14 |
Hb_010683_030 |
0.2568618412 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001266_150 |
0.2579556691 |
- |
- |
hypothetical protein JCGZ_14961 [Jatropha curcas] |
| 16 |
Hb_004447_050 |
0.2597725401 |
- |
- |
PREDICTED: high-light-induced protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000352_010 |
0.2611830796 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001221_200 |
0.2621085704 |
- |
- |
- |
| 19 |
Hb_000210_200 |
0.2622638616 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 20 |
Hb_001819_020 |
0.264846893 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |