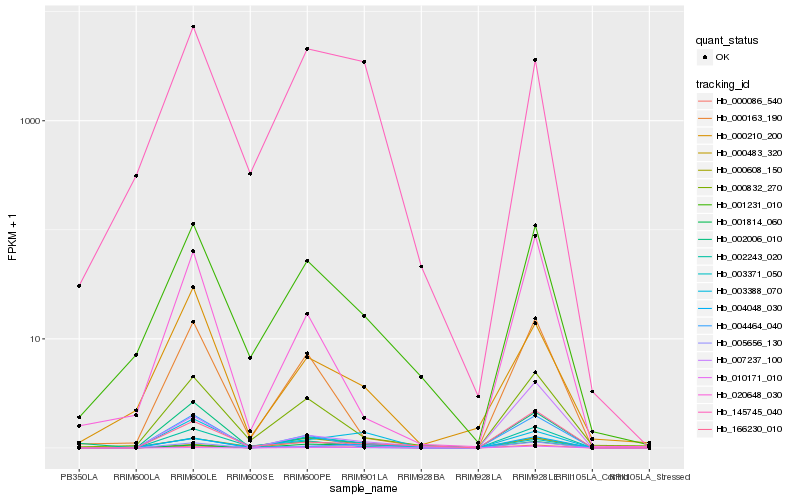
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005656_130 |
0.0 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 2 |
Hb_003388_070 |
0.1926138137 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 3 |
Hb_001231_010 |
0.228331285 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 4 |
Hb_003371_050 |
0.2323303199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010171_010 |
0.2344806721 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 6 |
Hb_000086_540 |
0.2361880723 |
- |
- |
- |
| 7 |
Hb_166230_010 |
0.236787583 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_004048_030 |
0.2410906079 |
- |
- |
hypothetical protein B456_009G371200, partial [Gossypium raimondii] |
| 9 |
Hb_145745_040 |
0.2518006459 |
- |
- |
Cell wall-associated hydrolase [Medicago truncatula] |
| 10 |
Hb_000832_270 |
0.2518326274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000483_320 |
0.2532693435 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002006_010 |
0.2655616277 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 13 |
Hb_004464_040 |
0.2697916822 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 14 |
Hb_000163_190 |
0.2700005327 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 15 |
Hb_000210_200 |
0.2726106429 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 16 |
Hb_001814_060 |
0.2728459725 |
- |
- |
PREDICTED: uncharacterized protein LOC105650637 [Jatropha curcas] |
| 17 |
Hb_000608_150 |
0.2735857942 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020648_030 |
0.2744934408 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 19 |
Hb_007237_100 |
0.2758753599 |
- |
- |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 20 |
Hb_002243_020 |
0.2763072829 |
- |
- |
PREDICTED: uncharacterized protein LOC105642097 isoform X1 [Jatropha curcas] |