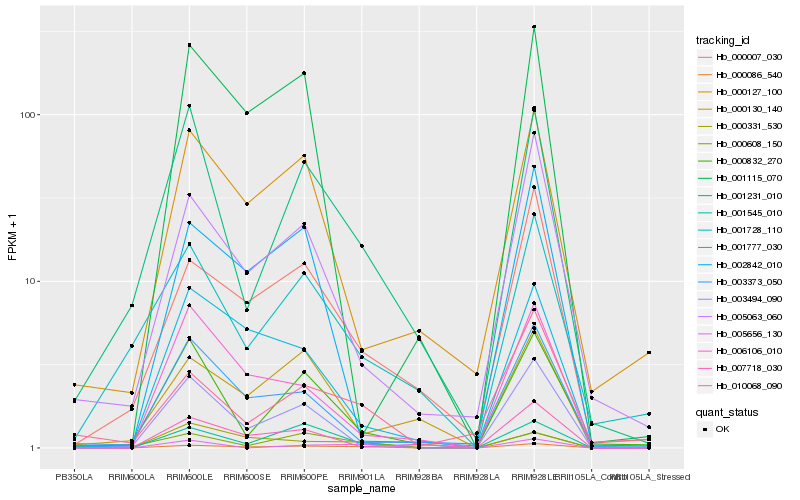
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_540 |
0.0 |
- |
- |
- |
| 2 |
Hb_001545_010 |
0.176203139 |
- |
- |
PREDICTED: serine/threonine-protein kinase TNNI3K-like [Cucumis sativus] |
| 3 |
Hb_000007_030 |
0.193716772 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000608_150 |
0.2058580488 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000331_530 |
0.214228281 |
- |
- |
PREDICTED: callose synthase 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001777_030 |
0.2285758237 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 7 |
Hb_005063_060 |
0.2292496185 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 8 |
Hb_001231_010 |
0.2333044563 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 9 |
Hb_005656_130 |
0.2361880723 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 10 |
Hb_010068_090 |
0.2389087154 |
- |
- |
hypothetical protein JCGZ_00885 [Jatropha curcas] |
| 11 |
Hb_002842_010 |
0.240542253 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 12 |
Hb_007718_030 |
0.2423648358 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_000127_100 |
0.2430008403 |
- |
- |
PREDICTED: myb-related transcription factor, partner of profilin-like [Populus euphratica] |
| 14 |
Hb_006106_010 |
0.2430467212 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000832_270 |
0.2441924566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003494_090 |
0.2443190478 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_003373_050 |
0.246079749 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 18 |
Hb_001728_110 |
0.2462171004 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 19 |
Hb_001115_070 |
0.2467840647 |
- |
- |
RecName: Full=CASP-like protein 2C1; Short=PtCASPL2C1 [Populus trichocarpa] |
| 20 |
Hb_000130_140 |
0.2470927708 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |