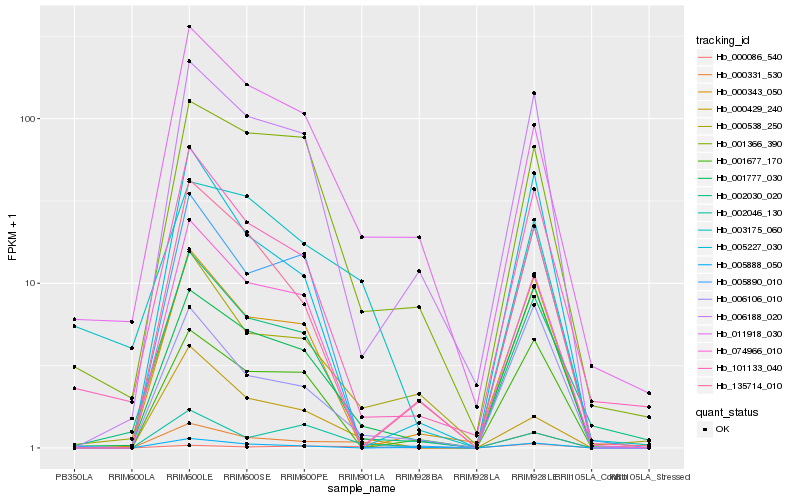
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_530 |
0.0 |
- |
- |
PREDICTED: callose synthase 10 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001777_030 |
0.1901090883 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 3 |
Hb_000538_250 |
0.2000095952 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 4 |
Hb_006106_010 |
0.2106135094 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000429_240 |
0.2138109268 |
- |
- |
PREDICTED: uncharacterized protein LOC105645185 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000086_540 |
0.214228281 |
- |
- |
- |
| 7 |
Hb_006188_020 |
0.2192291787 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 8 |
Hb_002030_020 |
0.2205173117 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 9 |
Hb_003175_060 |
0.2216752082 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 10 |
Hb_101133_040 |
0.2257013637 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_002046_130 |
0.2260635383 |
- |
- |
PREDICTED: UDP-sugar transporter sqv-7-like [Jatropha curcas] |
| 12 |
Hb_000343_050 |
0.2273478881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005227_030 |
0.2284212684 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 14 |
Hb_135714_010 |
0.2327862183 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 15 |
Hb_005890_010 |
0.2337576852 |
- |
- |
PREDICTED: root phototropism protein 2 [Jatropha curcas] |
| 16 |
Hb_005888_050 |
0.2368149286 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_011918_030 |
0.2386322042 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 18 |
Hb_074966_010 |
0.2398155743 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 19 |
Hb_001677_170 |
0.2418723579 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 20 |
Hb_001366_390 |
0.2423726561 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |