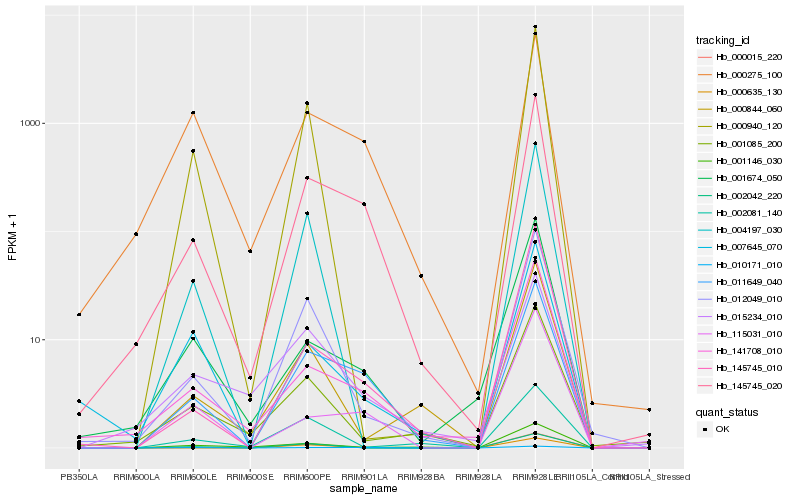
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011649_040 |
0.0 |
- |
- |
hypothetical protein CISIN_1g039733mg [Citrus sinensis] |
| 2 |
Hb_145745_020 |
0.0641471496 |
- |
- |
Cell wall-associated hydrolase, partial [Medicago truncatula] |
| 3 |
Hb_000275_100 |
0.1632999099 |
- |
- |
BnaC09g26910D [Brassica napus] |
| 4 |
Hb_010171_010 |
0.1701583472 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 5 |
Hb_141708_010 |
0.1724802546 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |
| 6 |
Hb_000015_220 |
0.178343368 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 7 |
Hb_012049_010 |
0.1811026858 |
- |
- |
hypothetical protein B456_002G055100 [Gossypium raimondii] |
| 8 |
Hb_115031_010 |
0.1891522659 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 9 |
Hb_000635_130 |
0.1904762159 |
- |
- |
hypothetical protein JCGZ_17952 [Jatropha curcas] |
| 10 |
Hb_015234_010 |
0.1921232624 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 11 |
Hb_001146_030 |
0.197049878 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_145745_010 |
0.1974831815 |
- |
- |
BnaUnng00830D [Brassica napus] |
| 13 |
Hb_001674_050 |
0.2052116854 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 14 |
Hb_007645_070 |
0.2092387159 |
- |
- |
photosystem II cytochrome b559 alpha subunit [Cuscuta obtusiflora] |
| 15 |
Hb_000844_060 |
0.2102563428 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 16 |
Hb_000940_120 |
0.2125970025 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 17 |
Hb_004197_030 |
0.2127123671 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 18 |
Hb_001085_200 |
0.2131344619 |
transcription factor |
TF Family: ARID |
PREDICTED: high mobility group B protein 15 [Jatropha curcas] |
| 19 |
Hb_002042_220 |
0.216690002 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_002081_140 |
0.2171559853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |