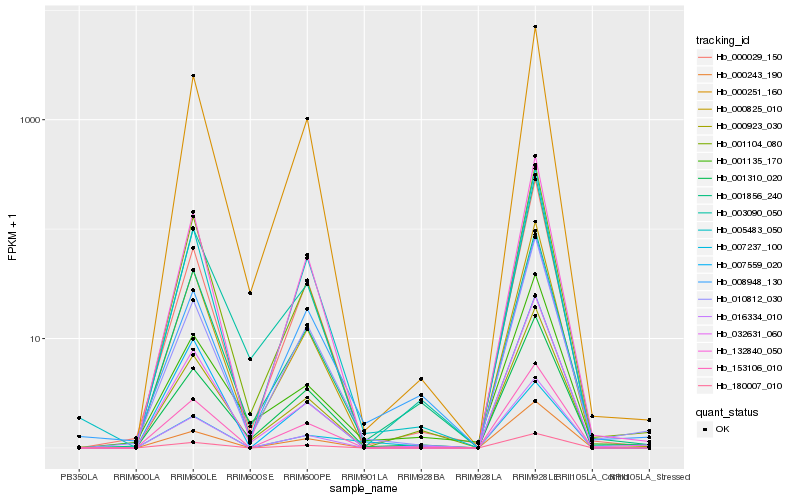
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007237_100 |
0.0 |
- |
- |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 2 |
Hb_001856_240 |
0.1396747716 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 3 |
Hb_005483_050 |
0.1402798808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000825_010 |
0.1418017164 |
- |
- |
PREDICTED: trans-cinnamate 4-monooxygenase [Jatropha curcas] |
| 5 |
Hb_001104_080 |
0.1419799942 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 6 |
Hb_016334_010 |
0.1424131341 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 7 |
Hb_008948_130 |
0.1427756702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_132840_050 |
0.1437951967 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 9 |
Hb_153106_010 |
0.1463175029 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 10 |
Hb_032631_060 |
0.1465714035 |
- |
- |
Myosin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_000029_150 |
0.1469672813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000923_030 |
0.1470997802 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 13 |
Hb_000251_160 |
0.1477049648 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 14 |
Hb_001310_020 |
0.148270566 |
- |
- |
PREDICTED: cyclin-D5-1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000243_190 |
0.1488271755 |
transcription factor |
TF Family: MIKC |
flowering-related B-class MADS-box protein [Vitis vinifera] |
| 16 |
Hb_010812_030 |
0.1490593926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_180007_010 |
0.1490931345 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 18 |
Hb_003090_050 |
0.1491316963 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 19 |
Hb_001135_170 |
0.1493624983 |
transcription factor |
TF Family: Orphans |
Two-component response regulator ARR2, putative [Ricinus communis] |
| 20 |
Hb_007559_020 |
0.1494154048 |
- |
- |
peptidylprolyl isomerase, putative [Ricinus communis] |