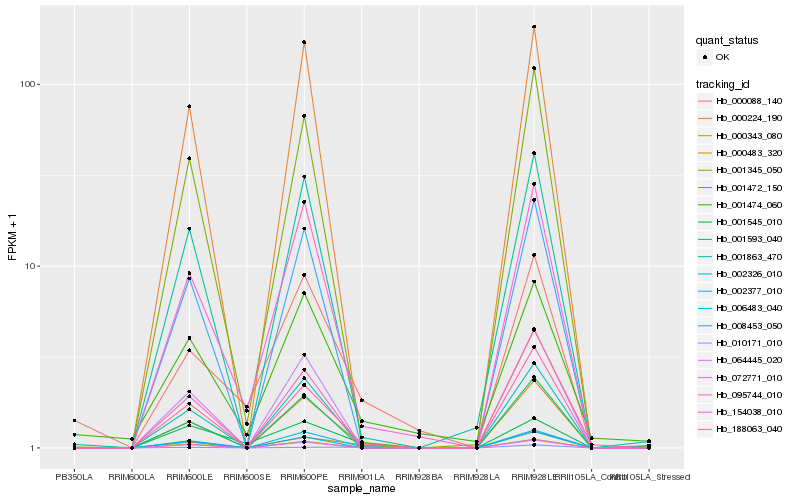
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000483_320 |
0.0 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002377_010 |
0.1334408218 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 2-like [Jatropha curcas] |
| 3 |
Hb_000343_080 |
0.1483872418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_188063_040 |
0.1967667356 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_095744_010 |
0.1975334348 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_001863_470 |
0.2009731109 |
- |
- |
PREDICTED: uncharacterized protein LOC105630613 [Jatropha curcas] |
| 7 |
Hb_001545_010 |
0.2015588853 |
- |
- |
PREDICTED: serine/threonine-protein kinase TNNI3K-like [Cucumis sativus] |
| 8 |
Hb_000088_140 |
0.2022126145 |
- |
- |
- |
| 9 |
Hb_010171_010 |
0.2045027762 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 10 |
Hb_154038_010 |
0.2056911549 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_006483_040 |
0.2057773566 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 12 |
Hb_001472_150 |
0.2058726868 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 13 |
Hb_064445_020 |
0.2063510098 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 14 |
Hb_008453_050 |
0.2068125851 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 15 |
Hb_002326_010 |
0.2079947073 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 16 |
Hb_001593_040 |
0.2080824269 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 17 |
Hb_072771_010 |
0.2092715642 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 18 |
Hb_001345_050 |
0.2094710292 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 19 |
Hb_001474_060 |
0.209660988 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 20 |
Hb_000224_190 |
0.2109523883 |
- |
- |
zinc finger protein, putative [Ricinus communis] |