| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
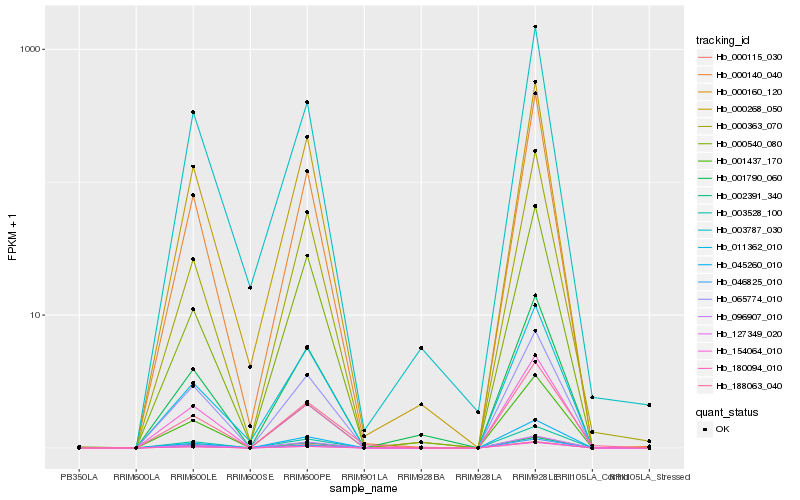
Hb_188063_040 |
0.0 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003528_100 |
0.0969879917 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 3 |
Hb_046825_010 |
0.1011423483 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 4 |
Hb_000160_120 |
0.1014569163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000363_070 |
0.101868276 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000268_050 |
0.1040618912 |
- |
- |
PREDICTED: tetrapyrrole-binding protein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_065774_010 |
0.1047923398 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001437_170 |
0.1048029265 |
- |
- |
hypothetical protein POPTR_0009s01390g [Populus trichocarpa] |
| 9 |
Hb_096907_010 |
0.1055907199 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 10 |
Hb_180094_010 |
0.108293755 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 11 |
Hb_154064_010 |
0.1092011051 |
- |
- |
PREDICTED: uncharacterized protein LOC105780682 [Gossypium raimondii] |
| 12 |
Hb_127349_020 |
0.1105312831 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 13 |
Hb_000140_040 |
0.1110290097 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 14 |
Hb_045260_010 |
0.1143347902 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 15 |
Hb_002391_340 |
0.1161344809 |
- |
- |
RNA polymerase II largest subunit [Liriodendron tulipifera] |
| 16 |
Hb_000540_080 |
0.1169707647 |
- |
- |
PREDICTED: laccase-17 [Jatropha curcas] |
| 17 |
Hb_011362_010 |
0.119123754 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 18 |
Hb_003787_030 |
0.1195144613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000115_030 |
0.1225973801 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 42 [Jatropha curcas] |
| 20 |
Hb_001790_060 |
0.123040087 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |