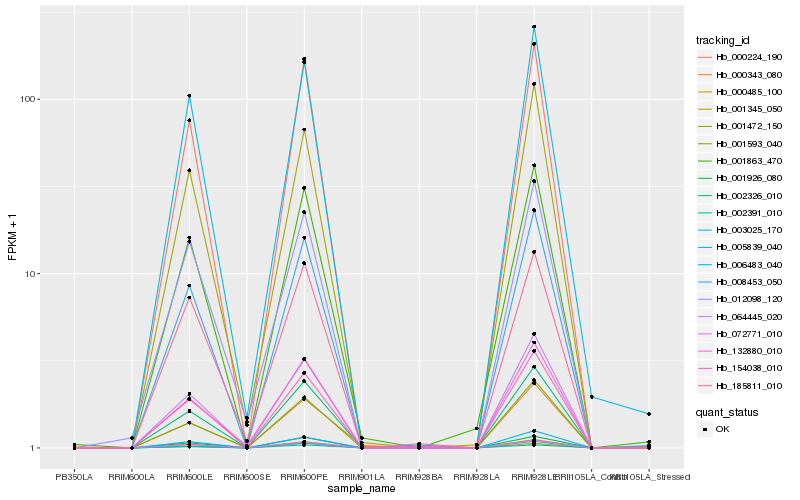
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630613 [Jatropha curcas] |
| 2 |
Hb_008453_050 |
0.0555908083 |
- |
- |
PREDICTED: peroxidase 64 [Jatropha curcas] |
| 3 |
Hb_072771_010 |
0.0601804494 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 4 |
Hb_002326_010 |
0.0603926727 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 5 |
Hb_001345_050 |
0.0605555594 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 6 |
Hb_000224_190 |
0.0606967449 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_154038_010 |
0.061877932 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_064445_020 |
0.0676557634 |
- |
- |
PREDICTED: serine carboxypeptidase-like 34 [Populus euphratica] |
| 9 |
Hb_006483_040 |
0.0707899183 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 10 |
Hb_001926_080 |
0.0713814852 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 11 |
Hb_001593_040 |
0.0726829507 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_005839_040 |
0.0757274334 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 13 |
Hb_002391_010 |
0.0777700481 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 14 |
Hb_001472_150 |
0.078920243 |
- |
- |
PREDICTED: cytochrome P450 86A7 [Jatropha curcas] |
| 15 |
Hb_000343_080 |
0.0847415513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000485_100 |
0.0920730243 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 17 |
Hb_185811_010 |
0.092184225 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_132880_010 |
0.0934969897 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 19 |
Hb_012098_120 |
0.0944199694 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 20 |
Hb_003025_170 |
0.0963184206 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |