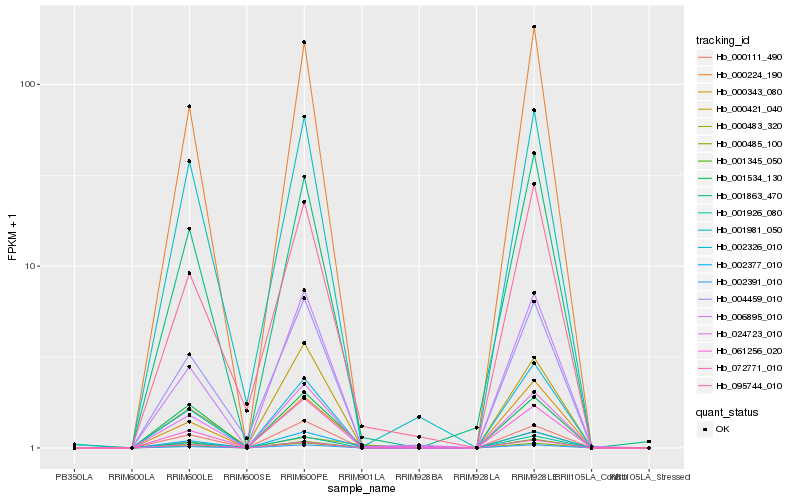
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002377_010 |
0.0 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 2-like [Jatropha curcas] |
| 2 |
Hb_000343_080 |
0.0901549179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002391_010 |
0.1183047161 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 4 |
Hb_001926_080 |
0.1190412555 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 5 |
Hb_000485_100 |
0.1197771812 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 6 |
Hb_001534_130 |
0.1202077264 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 7 |
Hb_006895_010 |
0.1221636361 |
- |
- |
PREDICTED: secologanin synthase-like [Citrus sinensis] |
| 8 |
Hb_000224_190 |
0.1227044525 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_095744_010 |
0.124629966 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_001863_470 |
0.1252065302 |
- |
- |
PREDICTED: uncharacterized protein LOC105630613 [Jatropha curcas] |
| 11 |
Hb_024723_010 |
0.1254795491 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 12 |
Hb_000111_490 |
0.1288850151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004459_010 |
0.1302874221 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 14 |
Hb_001345_050 |
0.1314097279 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 15 |
Hb_072771_010 |
0.1314150101 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 16 |
Hb_002326_010 |
0.1319485535 |
transcription factor |
TF Family: G2-like |
hypothetical protein RCOM_1592640 [Ricinus communis] |
| 17 |
Hb_000483_320 |
0.1334408218 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_061256_020 |
0.1359766991 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 19 |
Hb_000421_040 |
0.1379891951 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 20 |
Hb_001981_050 |
0.1385692079 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |