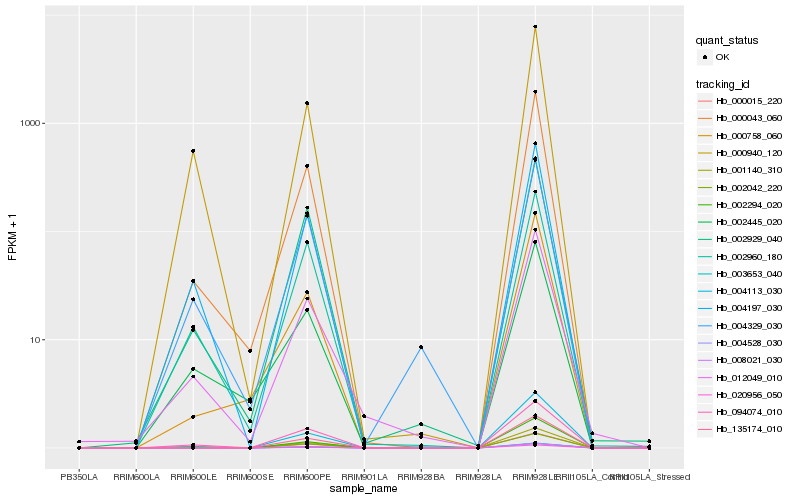
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000015_220 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 2 |
Hb_012049_010 |
0.0662298307 |
- |
- |
hypothetical protein B456_002G055100 [Gossypium raimondii] |
| 3 |
Hb_000043_060 |
0.0799501528 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 4 |
Hb_004197_030 |
0.0804849263 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 5 |
Hb_094074_010 |
0.081192668 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 6 |
Hb_002294_020 |
0.0903443085 |
- |
- |
PREDICTED: uncharacterized protein LOC105629874 [Jatropha curcas] |
| 7 |
Hb_002960_180 |
0.1020199476 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 8 |
Hb_002042_220 |
0.1023249157 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_000940_120 |
0.1034771714 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 10 |
Hb_002929_040 |
0.1079470432 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_004113_030 |
0.1144936684 |
- |
- |
- |
| 12 |
Hb_004329_030 |
0.1174066135 |
- |
- |
hypothetical protein JCGZ_17335 [Jatropha curcas] |
| 13 |
Hb_002445_020 |
0.1214005777 |
- |
- |
- |
| 14 |
Hb_000758_060 |
0.1214178387 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45960 [Jatropha curcas] |
| 15 |
Hb_135174_010 |
0.1232583658 |
- |
- |
Glycosyl hydrolase family protein 43 isoform 2 [Theobroma cacao] |
| 16 |
Hb_004528_030 |
0.1232584478 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 17 |
Hb_001140_310 |
0.1232584625 |
- |
- |
hypothetical protein AMTR_s00025p00025050 [Amborella trichopoda] |
| 18 |
Hb_008021_030 |
0.1232654583 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_003653_040 |
0.1233275586 |
- |
- |
hypothetical protein SORBIDRAFT_05g024215 [Sorghum bicolor] |
| 20 |
Hb_020956_050 |
0.1233347754 |
- |
- |
PREDICTED: uncharacterized protein LOC102670328 [Glycine max] |