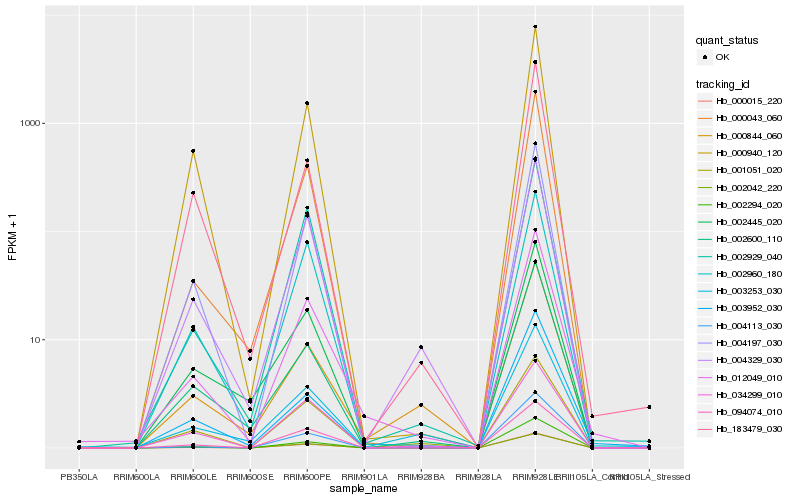
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012049_010 |
0.0 |
- |
- |
hypothetical protein B456_002G055100 [Gossypium raimondii] |
| 2 |
Hb_000015_220 |
0.0662298307 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os03g0620400-like [Jatropha curcas] |
| 3 |
Hb_004197_030 |
0.0934580231 |
- |
- |
hypothetical protein JCGZ_16263 [Jatropha curcas] |
| 4 |
Hb_000043_060 |
0.0955039679 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 5 |
Hb_094074_010 |
0.1004683538 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 6 |
Hb_004329_030 |
0.1056532238 |
- |
- |
hypothetical protein JCGZ_17335 [Jatropha curcas] |
| 7 |
Hb_000940_120 |
0.1066495236 |
- |
- |
PREDICTED: proline-rich protein 4-like isoform X2 [Pyrus x bretschneideri] |
| 8 |
Hb_002960_180 |
0.10678196 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 9 |
Hb_002042_220 |
0.1096675085 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_002294_020 |
0.1104393338 |
- |
- |
PREDICTED: uncharacterized protein LOC105629874 [Jatropha curcas] |
| 11 |
Hb_002600_110 |
0.1130496413 |
- |
- |
hypothetical protein POPTR_0004s10540g [Populus trichocarpa] |
| 12 |
Hb_002929_040 |
0.1132987203 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_003253_030 |
0.1177806116 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_002445_020 |
0.1178432259 |
- |
- |
- |
| 15 |
Hb_004113_030 |
0.1187452731 |
- |
- |
- |
| 16 |
Hb_001051_020 |
0.1202670518 |
- |
- |
hypothetical protein L484_008044 [Morus notabilis] |
| 17 |
Hb_000844_060 |
0.1216164074 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 18 |
Hb_003952_030 |
0.1217547028 |
- |
- |
hypothetical protein JCGZ_16620 [Jatropha curcas] |
| 19 |
Hb_034299_010 |
0.1241767888 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 20 |
Hb_183479_030 |
0.1270915574 |
- |
- |
Photosystem I reaction center subunit V, chloroplast precursor, putative [Ricinus communis] |