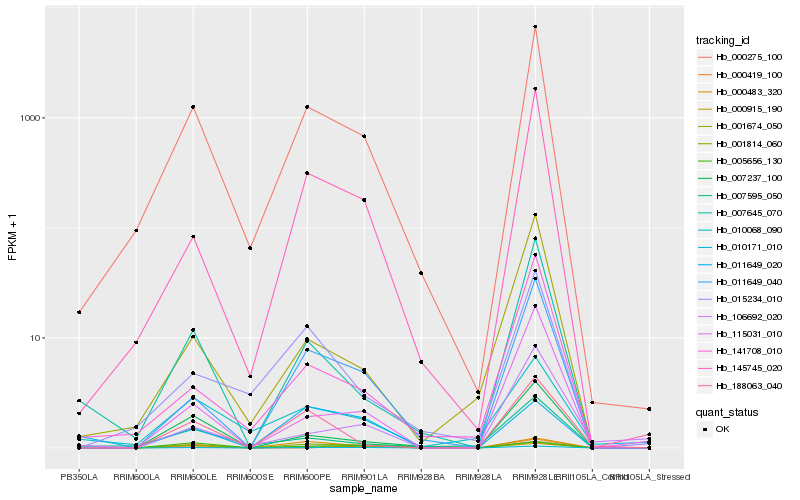
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010171_010 |
0.0 |
- |
- |
hypothetical protein VITISV_033646 [Vitis vinifera] |
| 2 |
Hb_000275_100 |
0.1574693422 |
- |
- |
BnaC09g26910D [Brassica napus] |
| 3 |
Hb_011649_040 |
0.1701583472 |
- |
- |
hypothetical protein CISIN_1g039733mg [Citrus sinensis] |
| 4 |
Hb_145745_020 |
0.1969742453 |
- |
- |
Cell wall-associated hydrolase, partial [Medicago truncatula] |
| 5 |
Hb_000483_320 |
0.2045027762 |
- |
- |
Major pollen allergen Ory s 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_007237_100 |
0.2316846934 |
- |
- |
DNA-directed RNA polymerase beta chain, putative [Ricinus communis] |
| 7 |
Hb_000915_190 |
0.2335805684 |
- |
- |
- |
| 8 |
Hb_005656_130 |
0.2344806721 |
- |
- |
PREDICTED: calmodulin-like protein 7 [Jatropha curcas] |
| 9 |
Hb_115031_010 |
0.2381144615 |
- |
- |
photosystem II P680 chlorophyll A apoprotein, partial (chloroplast) [Oenothera grandiflora] |
| 10 |
Hb_000419_100 |
0.2560620097 |
- |
- |
- |
| 11 |
Hb_007645_070 |
0.2591789745 |
- |
- |
photosystem II cytochrome b559 alpha subunit [Cuscuta obtusiflora] |
| 12 |
Hb_010068_090 |
0.2592236821 |
- |
- |
hypothetical protein JCGZ_00885 [Jatropha curcas] |
| 13 |
Hb_188063_040 |
0.2624362483 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001814_060 |
0.2653423797 |
- |
- |
PREDICTED: uncharacterized protein LOC105650637 [Jatropha curcas] |
| 15 |
Hb_015234_010 |
0.2667906074 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 16 |
Hb_106692_020 |
0.2671574135 |
- |
- |
NADH dehydrogenase subunit F, partial (chloroplast) [Hevea sp. Gillespie 4272] |
| 17 |
Hb_141708_010 |
0.2756477369 |
- |
- |
hypothetical protein EUTSA_v10011731mg [Eutrema salsugineum] |
| 18 |
Hb_011649_020 |
0.2778273332 |
- |
- |
PREDICTED: uncharacterized protein LOC104212904 isoform X1 [Nicotiana sylvestris] |
| 19 |
Hb_001674_050 |
0.2794580714 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 20 |
Hb_007595_050 |
0.2797426299 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |