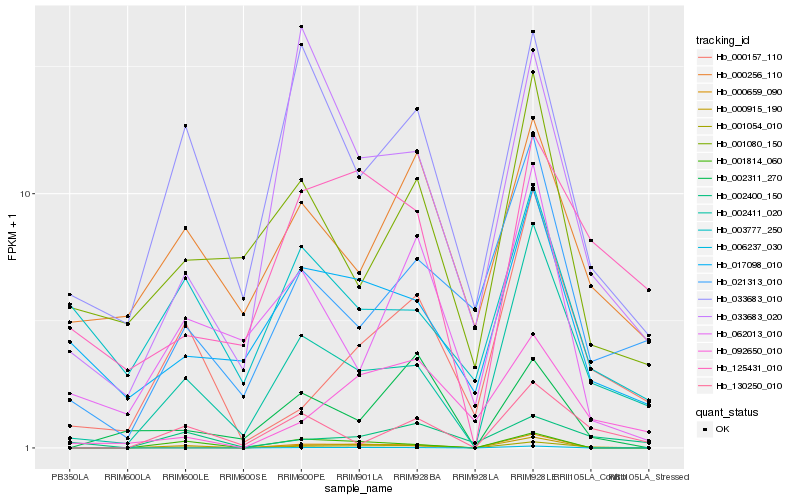
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001054_010 |
0.0 |
- |
- |
hypothetical protein VITISV_011091 [Vitis vinifera] |
| 2 |
Hb_002411_020 |
0.2178898134 |
- |
- |
hypothetical protein JCGZ_24577 [Jatropha curcas] |
| 3 |
Hb_000915_190 |
0.237102277 |
- |
- |
- |
| 4 |
Hb_000157_110 |
0.2651449561 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 5 |
Hb_017098_010 |
0.2665289256 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_125431_010 |
0.2676891513 |
- |
- |
- |
| 7 |
Hb_002400_150 |
0.2783531913 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 8 |
Hb_092650_010 |
0.2790446088 |
- |
- |
Uncharacterized protein TCM_017953 [Theobroma cacao] |
| 9 |
Hb_033683_010 |
0.2858104157 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 10 |
Hb_001814_060 |
0.2875435335 |
- |
- |
PREDICTED: uncharacterized protein LOC105650637 [Jatropha curcas] |
| 11 |
Hb_130250_010 |
0.2972078828 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 12 |
Hb_000659_090 |
0.2986464513 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 13 |
Hb_021313_010 |
0.300202714 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 14 |
Hb_062013_010 |
0.3008776632 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 15 |
Hb_001080_150 |
0.3036144481 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 16 |
Hb_006237_030 |
0.3042077723 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000256_110 |
0.3046873456 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_033683_020 |
0.3063138772 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 19 |
Hb_003777_250 |
0.3095189085 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 20 |
Hb_002311_270 |
0.3101038882 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |