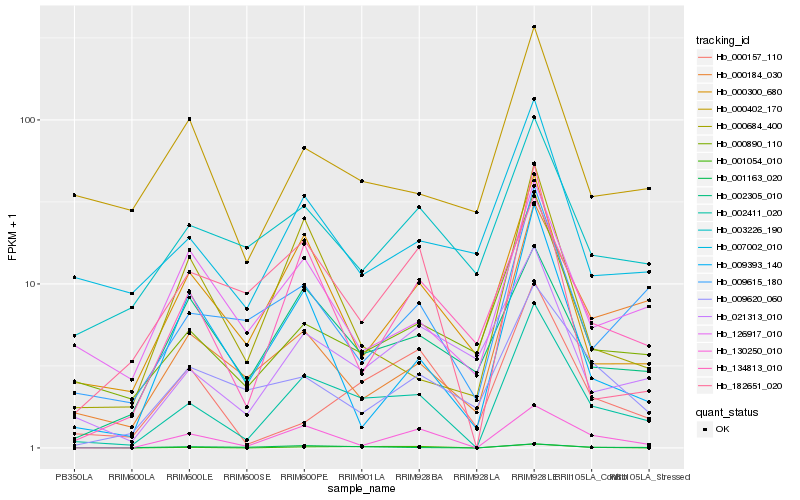
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_24577 [Jatropha curcas] |
| 2 |
Hb_130250_010 |
0.2034616154 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 3 |
Hb_000890_110 |
0.2061548618 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 4 |
Hb_134813_010 |
0.2166803592 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 5 |
Hb_000300_680 |
0.217127767 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001054_010 |
0.2178898134 |
- |
- |
hypothetical protein VITISV_011091 [Vitis vinifera] |
| 7 |
Hb_021313_010 |
0.2184349193 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 8 |
Hb_009615_180 |
0.2230136029 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 9 |
Hb_001163_020 |
0.2263048426 |
desease resistance |
Gene Name: ABC_trans_N |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_007002_010 |
0.2316964542 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 11 |
Hb_009393_140 |
0.233326864 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003226_190 |
0.2363127073 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 13 |
Hb_182651_020 |
0.2377137162 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000684_400 |
0.238038734 |
- |
- |
PREDICTED: uncharacterized protein LOC105642159 [Jatropha curcas] |
| 15 |
Hb_000157_110 |
0.2386821104 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |
| 16 |
Hb_002305_010 |
0.239977955 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_126917_010 |
0.2401170625 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 18 |
Hb_000184_030 |
0.2423814797 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000402_170 |
0.2439868174 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 20 |
Hb_009620_060 |
0.2443680239 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial-like [Jatropha curcas] |