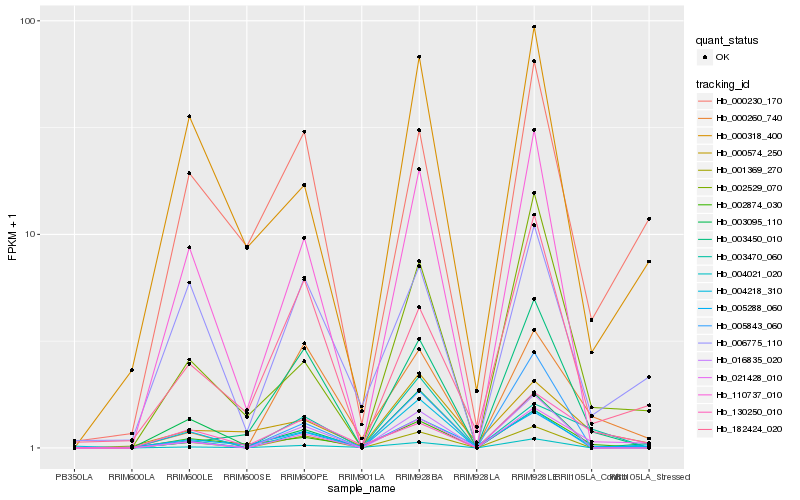
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021428_010 |
0.0 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 2 |
Hb_002529_070 |
0.1845457759 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 3 |
Hb_000260_740 |
0.205490426 |
- |
- |
- |
| 4 |
Hb_130250_010 |
0.2157302013 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 5 |
Hb_004021_020 |
0.2220939595 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 6 |
Hb_005288_060 |
0.2268525924 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_016835_020 |
0.2289825672 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_002874_030 |
0.2293661202 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 9 |
Hb_000230_170 |
0.2346945473 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 10 |
Hb_182424_020 |
0.2362691698 |
- |
- |
hypothetical protein POPTR_0006s24190g [Populus trichocarpa] |
| 11 |
Hb_000574_250 |
0.2385430293 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 12 |
Hb_006775_110 |
0.2423161739 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 13 |
Hb_004218_310 |
0.2456407944 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 14 |
Hb_001369_270 |
0.2457899007 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003470_060 |
0.2459769894 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 16 |
Hb_110737_010 |
0.2461526975 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 17 |
Hb_000318_400 |
0.2463073027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003450_010 |
0.2475379127 |
- |
- |
- |
| 19 |
Hb_003095_110 |
0.2476067789 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 20 |
Hb_005843_060 |
0.2536198412 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |