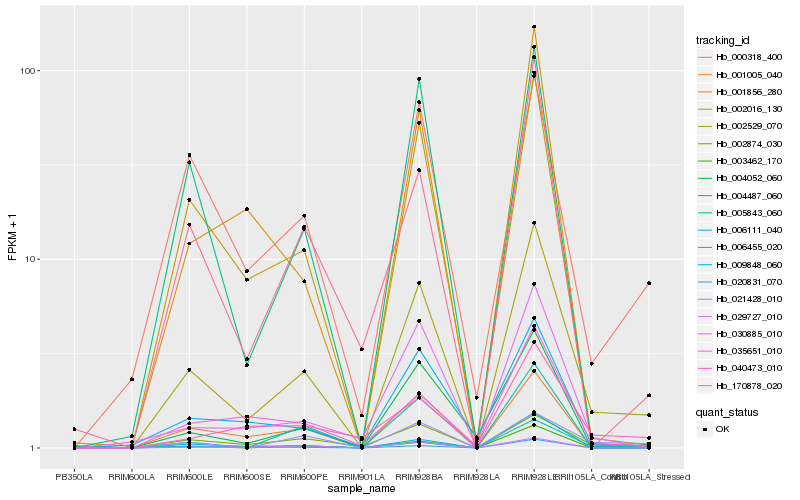
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002529_070 |
0.0 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 2 |
Hb_002016_130 |
0.1632648506 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 3 |
Hb_009848_060 |
0.1761269112 |
- |
- |
unknown [Glycine max] |
| 4 |
Hb_030885_010 |
0.1772304685 |
- |
- |
- |
| 5 |
Hb_001005_040 |
0.1788930722 |
- |
- |
- |
| 6 |
Hb_170878_020 |
0.180354215 |
- |
- |
- |
| 7 |
Hb_004052_060 |
0.1817273307 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_029727_010 |
0.1824039375 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_004487_060 |
0.1825351662 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_021428_010 |
0.1845457759 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 11 |
Hb_002874_030 |
0.1871500513 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 12 |
Hb_035651_010 |
0.1878060716 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 13 |
Hb_006111_040 |
0.188108791 |
- |
- |
PREDICTED: uncharacterized protein LOC103485923 [Cucumis melo] |
| 14 |
Hb_000318_400 |
0.1903949348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003462_170 |
0.1921708084 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_040473_010 |
0.1943906944 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 17 |
Hb_005843_060 |
0.1974236465 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 18 |
Hb_006455_020 |
0.1982920924 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001856_280 |
0.2004355648 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 20 |
Hb_020831_070 |
0.2010357349 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |