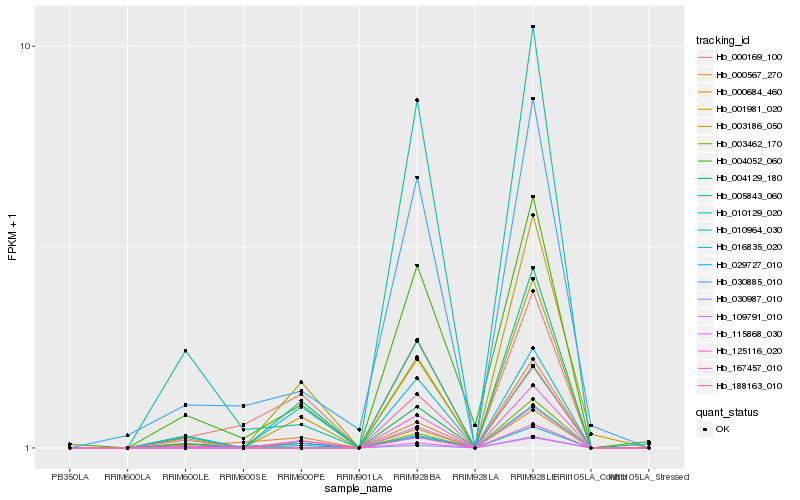
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_125116_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X5 [Populus euphratica] |
| 2 |
Hb_005843_060 |
0.0684793557 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 3 |
Hb_003462_170 |
0.1306281864 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_010964_030 |
0.1560760384 |
- |
- |
PREDICTED: uncharacterized protein LOC105647958 [Jatropha curcas] |
| 5 |
Hb_001981_020 |
0.1590227341 |
- |
- |
PREDICTED: xanthine dehydrogenase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_167457_010 |
0.1603177157 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 7 |
Hb_004052_060 |
0.1639210347 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_010129_020 |
0.167167247 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_003186_050 |
0.1679435487 |
- |
- |
- |
| 10 |
Hb_016835_020 |
0.1698813263 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_029727_010 |
0.1709841996 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_030987_010 |
0.1754147226 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_030885_010 |
0.1770688813 |
- |
- |
- |
| 14 |
Hb_000567_270 |
0.1813083877 |
- |
- |
hypothetical protein L484_017162 [Morus notabilis] |
| 15 |
Hb_004129_180 |
0.1817150322 |
- |
- |
hypothetical protein JCGZ_11090 [Jatropha curcas] |
| 16 |
Hb_000169_100 |
0.1933335029 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_188163_010 |
0.1955174191 |
- |
- |
hypothetical protein CISIN_1g048732mg [Citrus sinensis] |
| 18 |
Hb_109791_010 |
0.1992212031 |
- |
- |
- |
| 19 |
Hb_000684_460 |
0.199243889 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 20 |
Hb_115868_030 |
0.1992610446 |
- |
- |
Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial -like protein [Gossypium arboreum] |