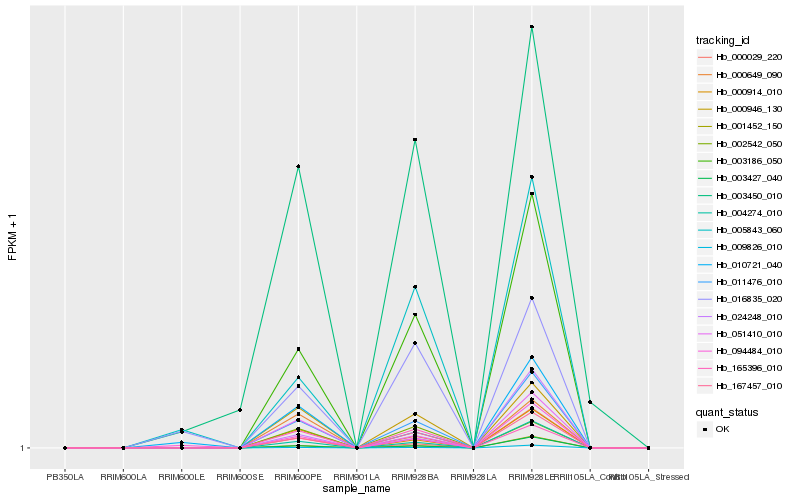
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011476_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_003186_050 |
0.0532830221 |
- |
- |
- |
| 3 |
Hb_167457_010 |
0.0636853585 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 4 |
Hb_004274_010 |
0.0825945924 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 5 |
Hb_001452_150 |
0.0992490109 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 6 |
Hb_000914_010 |
0.1061810331 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 7 |
Hb_003427_040 |
0.1072544812 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 8 |
Hb_000946_130 |
0.1087968293 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 9 |
Hb_051410_010 |
0.1100131239 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 10 |
Hb_009826_010 |
0.111672554 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_094484_010 |
0.15911032 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000649_090 |
0.1594300794 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000029_220 |
0.1634227856 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 14 |
Hb_005843_060 |
0.1701044517 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 15 |
Hb_165396_010 |
0.1790229536 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 16 |
Hb_010721_040 |
0.1793372651 |
transcription factor |
TF Family: M-type |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_024248_010 |
0.1794074117 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 18 |
Hb_002542_050 |
0.1795230925 |
- |
- |
- |
| 19 |
Hb_003450_010 |
0.1859863689 |
- |
- |
- |
| 20 |
Hb_016835_020 |
0.1886781688 |
- |
- |
unnamed protein product [Coffea canephora] |