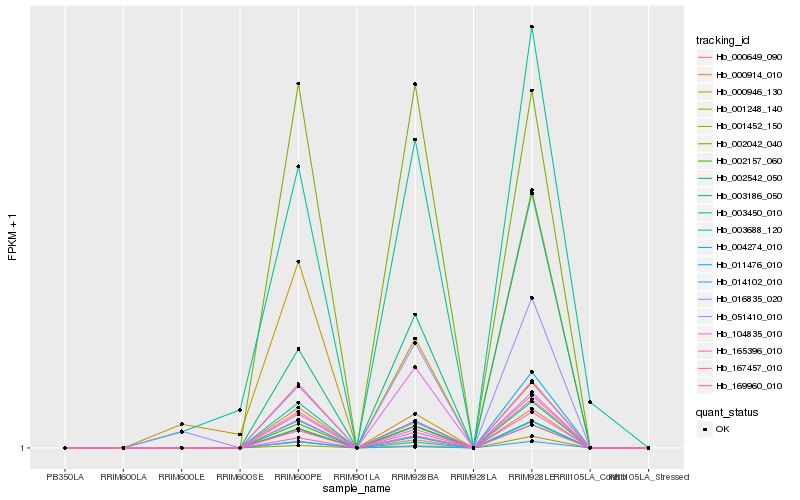
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000946_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 2 |
Hb_165396_010 |
0.0837078059 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 3 |
Hb_002542_050 |
0.0860938714 |
- |
- |
- |
| 4 |
Hb_011476_010 |
0.1087968293 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_003186_050 |
0.1243339094 |
- |
- |
- |
| 6 |
Hb_002157_060 |
0.1275152491 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 7 |
Hb_167457_010 |
0.1306225035 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 8 |
Hb_051410_010 |
0.1339915481 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 9 |
Hb_000649_090 |
0.1360120532 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_002042_040 |
0.1371778707 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 11 |
Hb_000914_010 |
0.1397681652 |
- |
- |
hypothetical protein POPTR_0025s00760g [Populus trichocarpa] |
| 12 |
Hb_169960_010 |
0.1550631957 |
- |
- |
- |
| 13 |
Hb_003450_010 |
0.1640979608 |
- |
- |
- |
| 14 |
Hb_001248_140 |
0.1692971095 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003688_120 |
0.1829142637 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_016835_020 |
0.1868089106 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_004274_010 |
0.1869374743 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 18 |
Hb_014102_010 |
0.1919781374 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_104835_010 |
0.1941770786 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 20 |
Hb_001452_150 |
0.2047533417 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |