| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
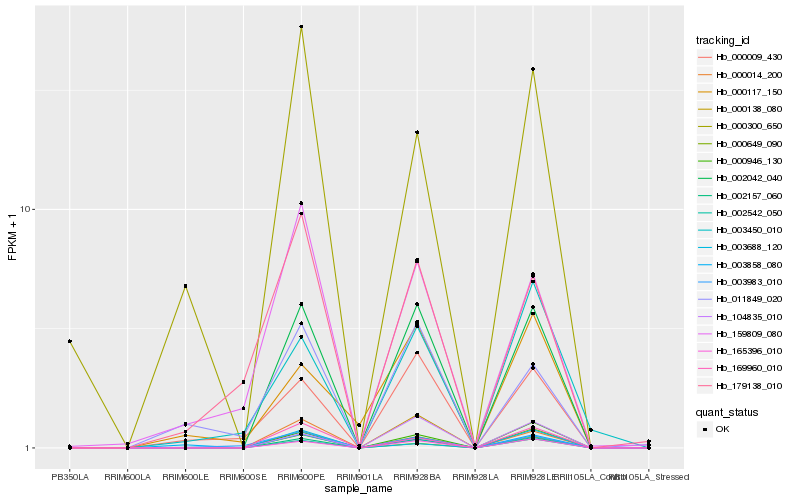
Hb_002157_060 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 2 |
Hb_002042_040 |
0.0176536864 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 3 |
Hb_165396_010 |
0.0625601558 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 4 |
Hb_002542_050 |
0.0649051695 |
- |
- |
- |
| 5 |
Hb_000946_130 |
0.1275152491 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 6 |
Hb_169960_010 |
0.1493755569 |
- |
- |
- |
| 7 |
Hb_000014_200 |
0.1613861592 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 8 |
Hb_104835_010 |
0.1644387437 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 9 |
Hb_000009_430 |
0.1799036405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003858_080 |
0.1947079219 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 11 |
Hb_003983_010 |
0.1957655831 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 12 |
Hb_159809_080 |
0.2024215994 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_179138_010 |
0.208263011 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_003450_010 |
0.2084501724 |
- |
- |
- |
| 15 |
Hb_011849_020 |
0.2168854133 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 16 |
Hb_000300_650 |
0.2185922671 |
- |
- |
PREDICTED: arabinogalactan peptide 13-like [Jatropha curcas] |
| 17 |
Hb_000138_080 |
0.2202525094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000649_090 |
0.2231137367 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000117_150 |
0.23041793 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 20 |
Hb_003688_120 |
0.2308159904 |
- |
- |
PREDICTED: uncharacterized protein LOC105350793 [Fragaria vesca subsp. vesca] |