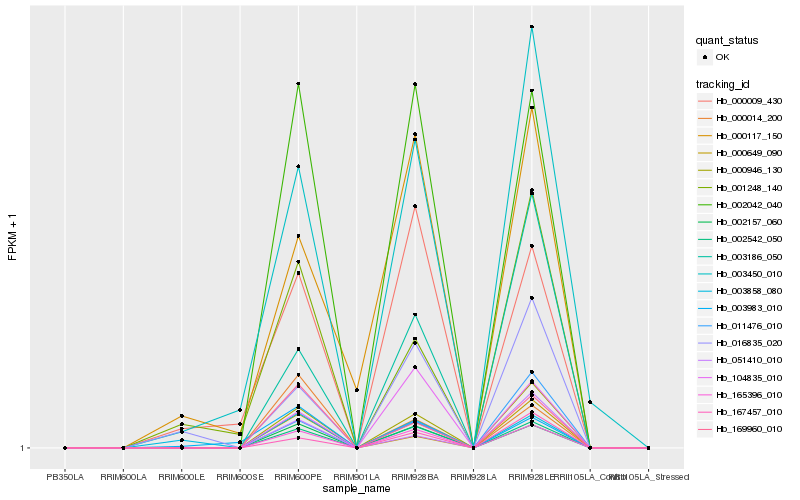
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165396_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 2 |
Hb_002542_050 |
0.0045965315 |
- |
- |
- |
| 3 |
Hb_002157_060 |
0.0625601558 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 4 |
Hb_002042_040 |
0.0637862407 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 5 |
Hb_000946_130 |
0.0837078059 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 6 |
Hb_104835_010 |
0.1318506227 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 7 |
Hb_169960_010 |
0.1733056403 |
- |
- |
- |
| 8 |
Hb_003186_050 |
0.1735425531 |
- |
- |
- |
| 9 |
Hb_003450_010 |
0.1751584854 |
- |
- |
- |
| 10 |
Hb_167457_010 |
0.1754102581 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 11 |
Hb_000009_430 |
0.1772373417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011476_010 |
0.1790229536 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 13 |
Hb_016835_020 |
0.1890234161 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000117_150 |
0.2026955865 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 15 |
Hb_003858_080 |
0.2046514676 |
- |
- |
Germin-like protein 2 [Theobroma cacao] |
| 16 |
Hb_000014_200 |
0.2065988577 |
- |
- |
PREDICTED: uncharacterized protein LOC105775355 [Gossypium raimondii] |
| 17 |
Hb_000649_090 |
0.2070158164 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_051410_010 |
0.2155368232 |
- |
- |
PREDICTED: uncharacterized protein LOC103401813 [Malus domestica] |
| 19 |
Hb_001248_140 |
0.2156588547 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_003983_010 |
0.2200539755 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |