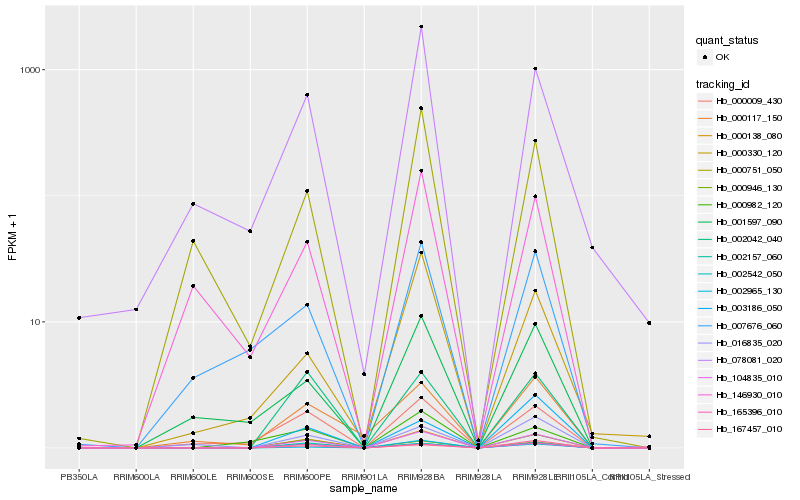
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_104835_010 |
0.0 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 2 |
Hb_002542_050 |
0.1273348995 |
- |
- |
- |
| 3 |
Hb_165396_010 |
0.1318506227 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 4 |
Hb_002042_040 |
0.150952097 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |
| 5 |
Hb_002157_060 |
0.1644387437 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 6 |
Hb_000009_430 |
0.1702933752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002965_130 |
0.1782416267 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 8 |
Hb_000138_080 |
0.1845700727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_016835_020 |
0.1920004304 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_078081_020 |
0.192116421 |
- |
- |
- |
| 11 |
Hb_000751_050 |
0.1927559457 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 12 |
Hb_001597_090 |
0.1937073582 |
- |
- |
- |
| 13 |
Hb_000946_130 |
0.1941770786 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 14 |
Hb_000117_150 |
0.1952935519 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 15 |
Hb_000330_120 |
0.2017716615 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 16 |
Hb_146930_010 |
0.2034235763 |
- |
- |
- |
| 17 |
Hb_000982_120 |
0.2036183026 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 18 |
Hb_167457_010 |
0.2050159912 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 19 |
Hb_007676_060 |
0.2085596605 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 20 |
Hb_003186_050 |
0.2099406015 |
- |
- |
- |