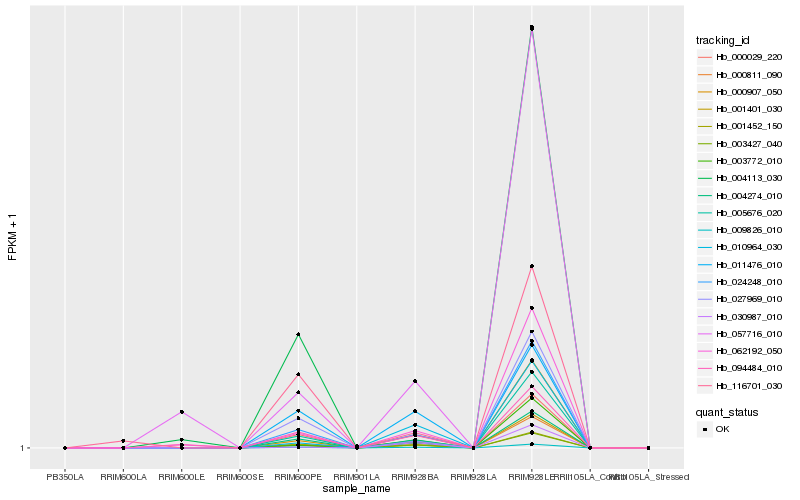
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024248_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 2 |
Hb_001401_030 |
0.0467726261 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 3 |
Hb_005676_020 |
0.0540864567 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 4 |
Hb_000029_220 |
0.0683339822 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 5 |
Hb_009826_010 |
0.0720596566 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_003427_040 |
0.0767782199 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 7 |
Hb_001452_150 |
0.0816693071 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 8 |
Hb_000907_050 |
0.0904467526 |
- |
- |
- |
| 9 |
Hb_004274_010 |
0.097622525 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 10 |
Hb_027969_010 |
0.1024909675 |
- |
- |
hypothetical protein JCGZ_24556 [Jatropha curcas] |
| 11 |
Hb_062192_050 |
0.1093644336 |
- |
- |
- |
| 12 |
Hb_003772_010 |
0.1258671128 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_094484_010 |
0.1628441301 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_057716_010 |
0.1635959008 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 15 |
Hb_010964_030 |
0.1703838049 |
- |
- |
PREDICTED: uncharacterized protein LOC105647958 [Jatropha curcas] |
| 16 |
Hb_000811_090 |
0.1762874039 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 17 |
Hb_030987_010 |
0.1783847732 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_004113_030 |
0.1792314855 |
- |
- |
- |
| 19 |
Hb_116701_030 |
0.1793733723 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 20 |
Hb_011476_010 |
0.1794074117 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |