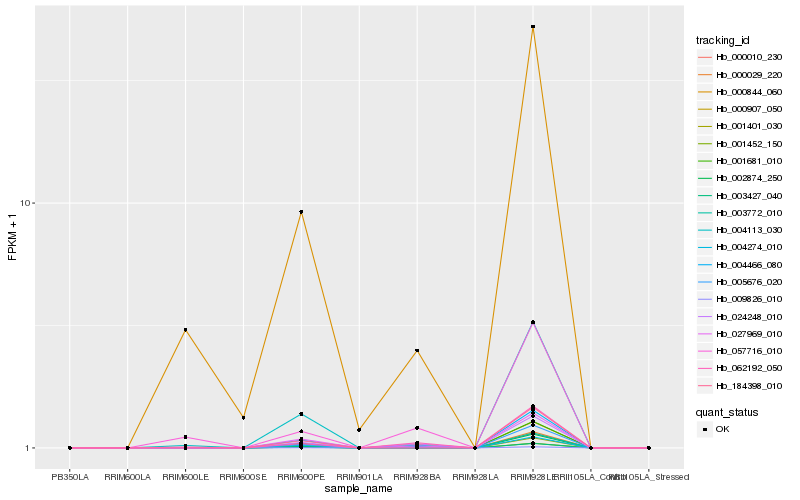
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005676_020 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 2 |
Hb_024248_010 |
0.0540864567 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 3 |
Hb_001401_030 |
0.0572155546 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 4 |
Hb_027969_010 |
0.0747484334 |
- |
- |
hypothetical protein JCGZ_24556 [Jatropha curcas] |
| 5 |
Hb_000029_220 |
0.0817009355 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 6 |
Hb_000907_050 |
0.1159027162 |
- |
- |
- |
| 7 |
Hb_062192_050 |
0.118074634 |
- |
- |
- |
| 8 |
Hb_009826_010 |
0.1240991521 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_003772_010 |
0.1280132725 |
- |
- |
polyprotein [Oryza australiensis] |
| 10 |
Hb_003427_040 |
0.1286904823 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 11 |
Hb_001452_150 |
0.131840579 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 12 |
Hb_004113_030 |
0.1358562649 |
- |
- |
- |
| 13 |
Hb_004274_010 |
0.1455719794 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 14 |
Hb_000844_060 |
0.1573723503 |
- |
- |
PREDICTED: uncharacterized protein LOC105633706 [Jatropha curcas] |
| 15 |
Hb_057716_010 |
0.1649450666 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 16 |
Hb_184398_010 |
0.166487393 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 17 |
Hb_001681_010 |
0.1667417333 |
- |
- |
PREDICTED: cytochrome P450 81E8-like isoform X1 [Populus euphratica] |
| 18 |
Hb_004466_080 |
0.1692637598 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 19 |
Hb_000010_230 |
0.1708963078 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 20 |
Hb_002874_250 |
0.1713081552 |
- |
- |
- |