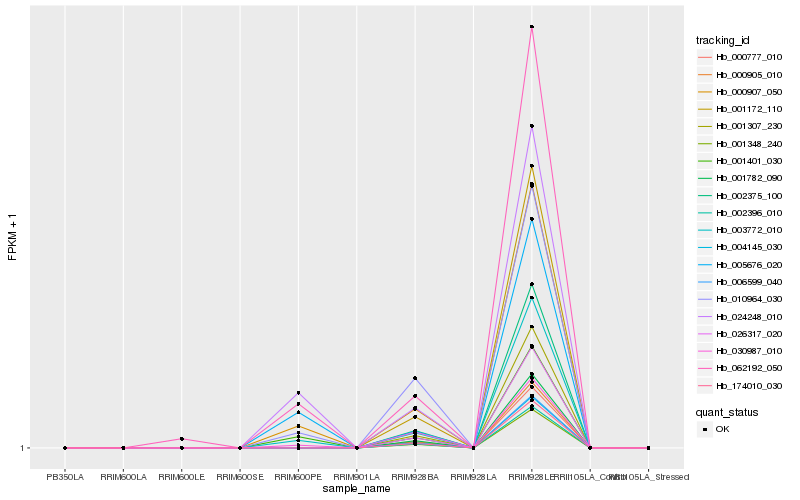
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003772_010 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_000907_050 |
0.0576181992 |
- |
- |
- |
| 3 |
Hb_001401_030 |
0.0814176203 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 4 |
Hb_062192_050 |
0.1049058613 |
- |
- |
- |
| 5 |
Hb_030987_010 |
0.1200976306 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_024248_010 |
0.1258671128 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 7 |
Hb_005676_020 |
0.1280132725 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 8 |
Hb_010964_030 |
0.1322917836 |
- |
- |
PREDICTED: uncharacterized protein LOC105647958 [Jatropha curcas] |
| 9 |
Hb_001782_090 |
0.1448230211 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62910-like [Vitis vinifera] |
| 10 |
Hb_000905_010 |
0.1448244286 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 11 |
Hb_000777_010 |
0.1448258896 |
- |
- |
PREDICTED: uncharacterized protein LOC105766834 [Gossypium raimondii] |
| 12 |
Hb_026317_020 |
0.1448291275 |
- |
- |
hypothetical protein VITISV_035911 [Vitis vinifera] |
| 13 |
Hb_006599_040 |
0.1448298083 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_004145_030 |
0.1448304317 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 15 |
Hb_174010_030 |
0.1448325152 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_002396_010 |
0.1448366857 |
- |
- |
PREDICTED: uncharacterized protein LOC105767002 [Gossypium raimondii] |
| 17 |
Hb_001348_240 |
0.1448389446 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 18 |
Hb_001307_230 |
0.1448457395 |
- |
- |
PREDICTED: interferon-related developmental regulator 1-like [Jatropha curcas] |
| 19 |
Hb_001172_110 |
0.1448895506 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 20 |
Hb_002375_100 |
0.144918735 |
- |
- |
PREDICTED: uncharacterized protein LOC104878944 [Vitis vinifera] |