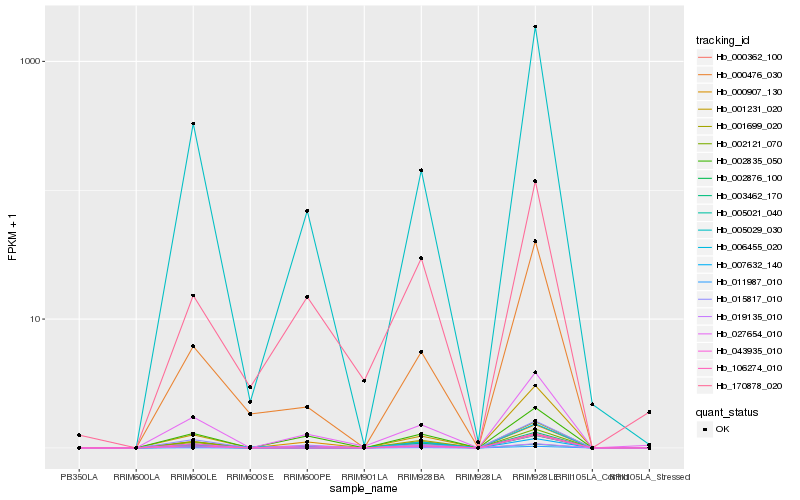
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001699_020 |
0.0 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 2 |
Hb_002876_100 |
0.0864738407 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 3 |
Hb_003462_170 |
0.1207361216 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_007632_140 |
0.1355774686 |
- |
- |
- |
| 5 |
Hb_027654_010 |
0.1376705151 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 6 |
Hb_019135_010 |
0.138137071 |
- |
- |
- |
| 7 |
Hb_015817_010 |
0.1381555503 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 8 |
Hb_006455_020 |
0.141248746 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000362_100 |
0.1419470996 |
- |
- |
- |
| 10 |
Hb_000476_030 |
0.1421113152 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 11 |
Hb_005029_030 |
0.1506474363 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 12 |
Hb_005021_040 |
0.150987185 |
- |
- |
PREDICTED: uncharacterized protein LOC103848240 [Brassica rapa] |
| 13 |
Hb_002121_070 |
0.1515105103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011987_010 |
0.1524226166 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 15 |
Hb_106274_010 |
0.1570042849 |
- |
- |
hypothetical protein VITISV_008825 [Vitis vinifera] |
| 16 |
Hb_170878_020 |
0.1579434152 |
- |
- |
- |
| 17 |
Hb_000907_130 |
0.1603510509 |
- |
- |
- |
| 18 |
Hb_043935_010 |
0.1615717452 |
- |
- |
Receptor protein kinase-like protein [Medicago truncatula] |
| 19 |
Hb_002835_050 |
0.1666516148 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 20 |
Hb_001231_020 |
0.168492581 |
- |
- |
- |