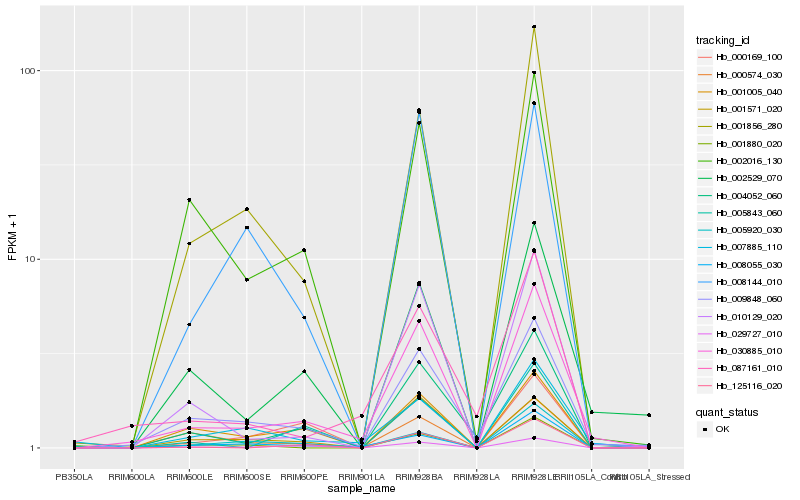
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030885_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_009848_060 |
0.1479716313 |
- |
- |
unknown [Glycine max] |
| 3 |
Hb_010129_020 |
0.1652967096 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001856_280 |
0.1752197478 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 5 |
Hb_029727_010 |
0.1760786308 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_125116_020 |
0.1770688813 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X5 [Populus euphratica] |
| 7 |
Hb_002529_070 |
0.1772304685 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 8 |
Hb_007885_110 |
0.1836799535 |
- |
- |
PREDICTED: uncharacterized protein LOC104109376 [Nicotiana tomentosiformis] |
| 9 |
Hb_001571_020 |
0.1859793589 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850-like [Citrus sinensis] |
| 10 |
Hb_004052_060 |
0.1873195722 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_008055_030 |
0.1889673882 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 12 |
Hb_087161_010 |
0.1891829362 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_008144_010 |
0.1923207139 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 14 |
Hb_002016_130 |
0.1933311786 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 15 |
Hb_000574_030 |
0.1934936565 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 16 |
Hb_000169_100 |
0.1980518115 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_005920_030 |
0.198897087 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 18 |
Hb_001005_040 |
0.1989719872 |
- |
- |
- |
| 19 |
Hb_005843_060 |
0.2013322147 |
- |
- |
PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X3 [Nelumbo nucifera] |
| 20 |
Hb_001880_020 |
0.20251421 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |