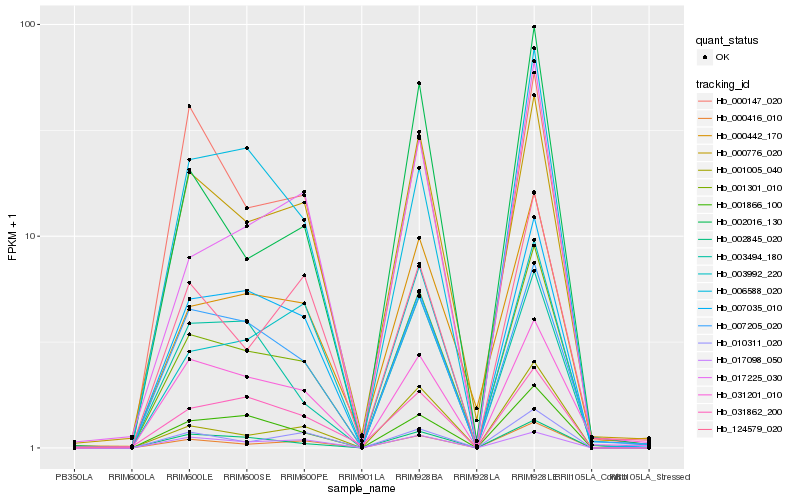
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_010 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000776_020 |
0.0754100407 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 3 |
Hb_000416_010 |
0.0944323858 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 4 |
Hb_002845_020 |
0.0984159732 |
- |
- |
hypothetical protein POPTR_0012s14920g [Populus trichocarpa] |
| 5 |
Hb_000442_170 |
0.1067287207 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_001866_100 |
0.1150797028 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 7 |
Hb_007205_020 |
0.1178342212 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 8 |
Hb_001005_040 |
0.1257060279 |
- |
- |
- |
| 9 |
Hb_007035_010 |
0.1267402105 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010311_020 |
0.1312516745 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_031201_010 |
0.1365745958 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002016_130 |
0.1369753902 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 13 |
Hb_003992_220 |
0.1384875632 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 14 |
Hb_124579_020 |
0.1394194851 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 15 |
Hb_017225_030 |
0.1406458065 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 16 |
Hb_031862_200 |
0.1431102033 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 17 |
Hb_003494_180 |
0.1434384416 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 18 |
Hb_006588_020 |
0.145366361 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000147_020 |
0.1456047902 |
- |
- |
PREDICTED: uncharacterized protein LOC105638303 [Jatropha curcas] |
| 20 |
Hb_017098_050 |
0.1461862589 |
- |
- |
hypothetical protein POPTR_0001s03540g [Populus trichocarpa] |