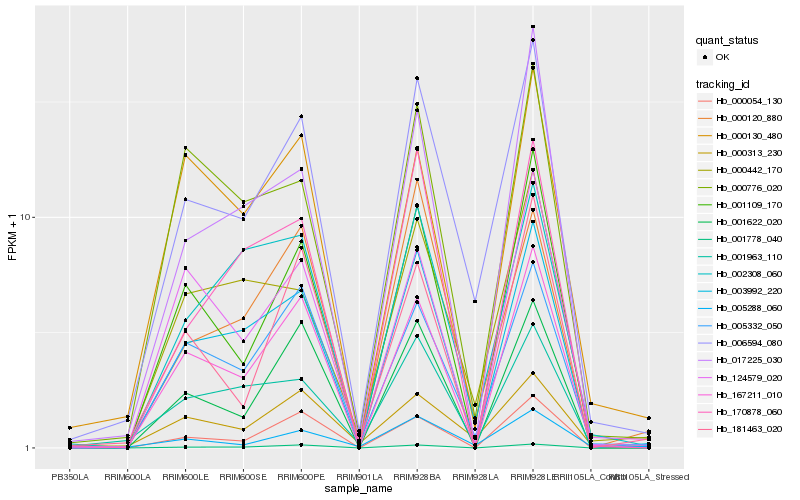
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006594_080 |
0.0 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_005332_050 |
0.1313818972 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 3 |
Hb_003992_220 |
0.131626267 |
- |
- |
s-receptor kinase, putative [Ricinus communis] |
| 4 |
Hb_167211_010 |
0.1318500877 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000442_170 |
0.1348705265 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 6 |
Hb_000313_230 |
0.1351628917 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 7 |
Hb_005288_060 |
0.1354538248 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_001109_170 |
0.142237236 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001963_110 |
0.1426565518 |
- |
- |
- |
| 10 |
Hb_001778_040 |
0.149968973 |
- |
- |
hypothetical protein VITISV_021035 [Vitis vinifera] |
| 11 |
Hb_002308_060 |
0.1506397817 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 12 |
Hb_000054_130 |
0.1523807904 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 13 |
Hb_001622_020 |
0.1524790213 |
- |
- |
- |
| 14 |
Hb_000120_880 |
0.156719894 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 15 |
Hb_000776_020 |
0.160490754 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 16 |
Hb_017225_030 |
0.1614893573 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 17 |
Hb_170878_060 |
0.1632857261 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 18 |
Hb_124579_020 |
0.1646193697 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 19 |
Hb_181463_020 |
0.1687026262 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 20 |
Hb_000130_480 |
0.1697092784 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |