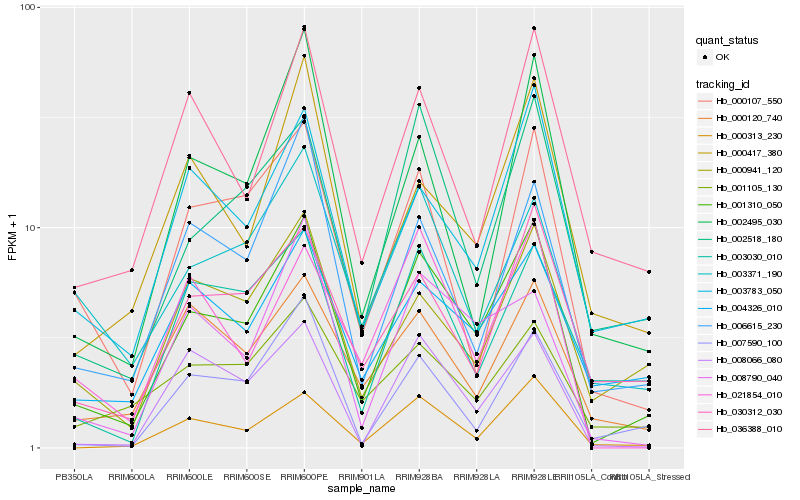
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001310_050 |
0.0 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 2 |
Hb_000313_230 |
0.1440797606 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 3 |
Hb_002518_180 |
0.1448488834 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 4 |
Hb_003783_050 |
0.1509318458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_030312_030 |
0.1580291821 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 6 |
Hb_036388_010 |
0.1606799313 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 7 |
Hb_004326_010 |
0.1608293496 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_000941_120 |
0.1621416683 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 9 |
Hb_008066_080 |
0.1621933391 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 10 |
Hb_000107_550 |
0.1622620122 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 11 |
Hb_008790_040 |
0.1656696251 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 12 |
Hb_000120_740 |
0.1667311642 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002495_030 |
0.1675634164 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006615_230 |
0.168436334 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 15 |
Hb_007590_100 |
0.1687565497 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 16 |
Hb_001105_130 |
0.1697470259 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 17 |
Hb_021854_010 |
0.1698227333 |
- |
- |
- |
| 18 |
Hb_000417_380 |
0.1722159097 |
- |
- |
SecY protein transport family protein [Theobroma cacao] |
| 19 |
Hb_003030_010 |
0.1723183481 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 20 |
Hb_003371_190 |
0.1733805544 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |