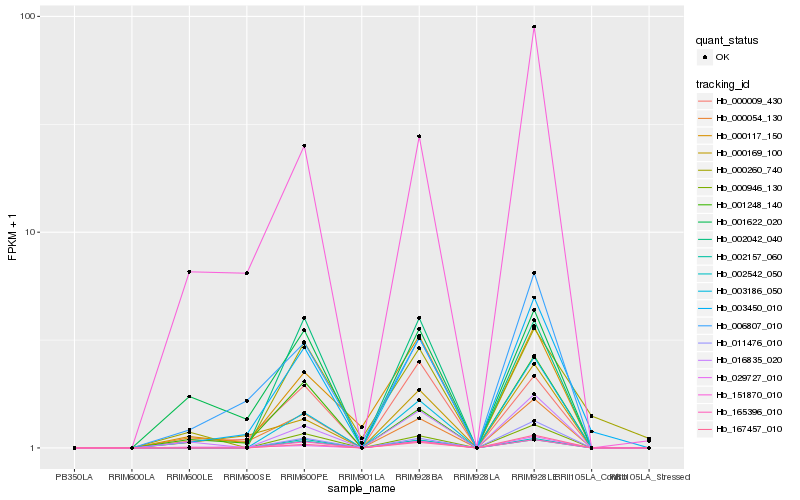
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003450_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000946_130 |
0.1640979608 |
- |
- |
PREDICTED: uncharacterized protein LOC104602430 [Nelumbo nucifera] |
| 3 |
Hb_006807_010 |
0.1691132411 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 4 |
Hb_002542_050 |
0.1750006433 |
- |
- |
- |
| 5 |
Hb_165396_010 |
0.1751584854 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 6 |
Hb_000169_100 |
0.1753096335 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_001248_140 |
0.1762189679 |
- |
- |
Thylakoid lumenal 19 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_003186_050 |
0.181951959 |
- |
- |
- |
| 9 |
Hb_167457_010 |
0.1834868532 |
- |
- |
hypothetical protein glysoja_045622, partial [Glycine soja] |
| 10 |
Hb_016835_020 |
0.1848759381 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_011476_010 |
0.1859863689 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 12 |
Hb_151870_010 |
0.1903503537 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_000009_430 |
0.1912432777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000054_130 |
0.1956349314 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 15 |
Hb_029727_010 |
0.1962547767 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_000117_150 |
0.2009308154 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 17 |
Hb_000260_740 |
0.2021041793 |
- |
- |
- |
| 18 |
Hb_002157_060 |
0.2084501724 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 19 |
Hb_001622_020 |
0.2104364576 |
- |
- |
- |
| 20 |
Hb_002042_040 |
0.2109367384 |
- |
- |
hypothetical protein POPTR_0013s02440g [Populus trichocarpa] |