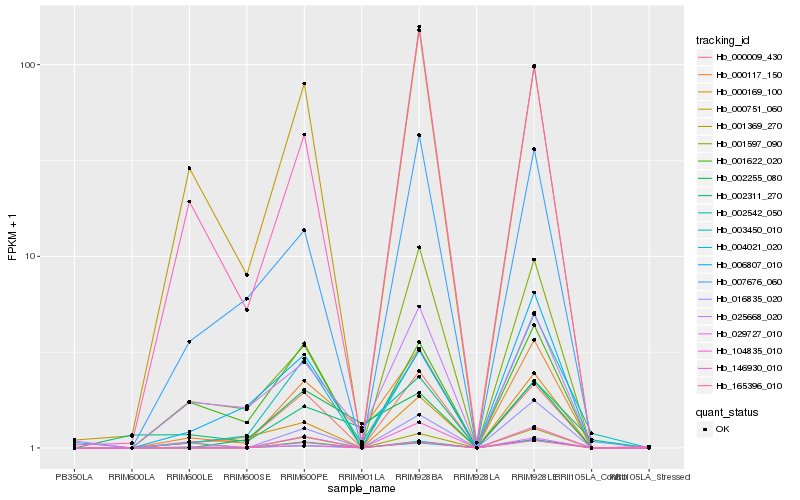
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_150 |
0.0 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 2 |
Hb_016835_020 |
0.1725563736 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_025668_020 |
0.1731811305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000009_430 |
0.1755094971 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002255_080 |
0.1886528486 |
- |
- |
- |
| 6 |
Hb_001597_090 |
0.189751479 |
- |
- |
- |
| 7 |
Hb_104835_010 |
0.1952935519 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 8 |
Hb_007676_060 |
0.1988786701 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 9 |
Hb_000169_100 |
0.2002086558 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_002542_050 |
0.2009303939 |
- |
- |
- |
| 11 |
Hb_003450_010 |
0.2009308154 |
- |
- |
- |
| 12 |
Hb_004021_020 |
0.2025823821 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 13 |
Hb_165396_010 |
0.2026955865 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 14 |
Hb_146930_010 |
0.205579713 |
- |
- |
- |
| 15 |
Hb_029727_010 |
0.2074485072 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_006807_010 |
0.2082267082 |
- |
- |
hypothetical protein JCGZ_20881 [Jatropha curcas] |
| 17 |
Hb_001369_270 |
0.2122214797 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000751_060 |
0.2144640925 |
- |
- |
PREDICTED: extensin-2-like, partial [Sesamum indicum] |
| 19 |
Hb_002311_270 |
0.216915488 |
- |
- |
PREDICTED: WD repeat-containing protein 43-like [Malus domestica] |
| 20 |
Hb_001622_020 |
0.2192218073 |
- |
- |
- |