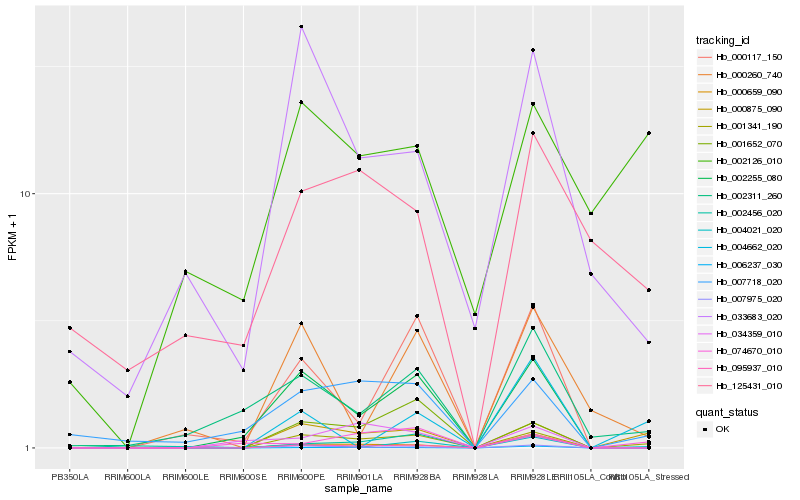
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001341_190 |
0.0 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000875_090 |
0.1109481996 |
- |
- |
hypothetical protein VITISV_005326 [Vitis vinifera] |
| 3 |
Hb_007975_020 |
0.2431128901 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_002255_080 |
0.2438895459 |
- |
- |
- |
| 5 |
Hb_001652_070 |
0.2558106242 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 6 |
Hb_007718_020 |
0.266926466 |
- |
- |
PREDICTED: receptor-like protein 12 [Camelina sativa] |
| 7 |
Hb_000659_090 |
0.3017178492 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 8 |
Hb_002311_260 |
0.3027981555 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 9 |
Hb_000117_150 |
0.3092682932 |
- |
- |
hypothetical protein RCOM_0079810 [Ricinus communis] |
| 10 |
Hb_004021_020 |
0.3099767795 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 11 |
Hb_074670_010 |
0.3118418835 |
- |
- |
hypothetical protein VITISV_015625 [Vitis vinifera] |
| 12 |
Hb_033683_020 |
0.3128419547 |
- |
- |
hypothetical protein POPTR_1072s00200g [Populus trichocarpa] |
| 13 |
Hb_006237_030 |
0.3281498376 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_002456_020 |
0.3288855491 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 15 |
Hb_000260_740 |
0.3305865032 |
- |
- |
- |
| 16 |
Hb_002126_010 |
0.3328361368 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 17 |
Hb_095937_010 |
0.3375069028 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_034359_010 |
0.3410467025 |
- |
- |
- |
| 19 |
Hb_004662_020 |
0.3461481583 |
- |
- |
40S ribosomal protein S9, putative [Ricinus communis] |
| 20 |
Hb_125431_010 |
0.3463346662 |
- |
- |
- |