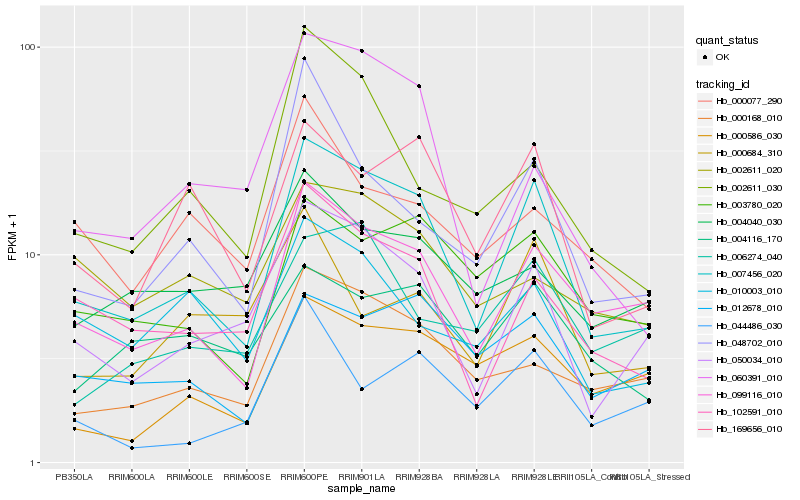
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007456_020 |
0.0 |
- |
- |
unknown [Lotus japonicus] |
| 2 |
Hb_099116_010 |
0.1186074558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_050034_010 |
0.1266012833 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 4 |
Hb_102591_010 |
0.1467607014 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 5 |
Hb_169656_010 |
0.1545829791 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 6 |
Hb_003780_020 |
0.1651193994 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |
| 7 |
Hb_000168_010 |
0.1656519553 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 8 |
Hb_012678_010 |
0.1750562482 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 9 |
Hb_048702_010 |
0.1779175833 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_060391_010 |
0.1789233476 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_004116_170 |
0.182772661 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000586_030 |
0.1829871783 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 13 |
Hb_002611_030 |
0.1860819564 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 14 |
Hb_006274_040 |
0.186585816 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 15 |
Hb_010003_010 |
0.1871051191 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004040_030 |
0.1977308819 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 17 |
Hb_044486_030 |
0.1988341216 |
- |
- |
DMI1, partial [Cycas revoluta] |
| 18 |
Hb_000077_290 |
0.1991578916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000684_310 |
0.2000141681 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 20 |
Hb_002611_020 |
0.2006470006 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |