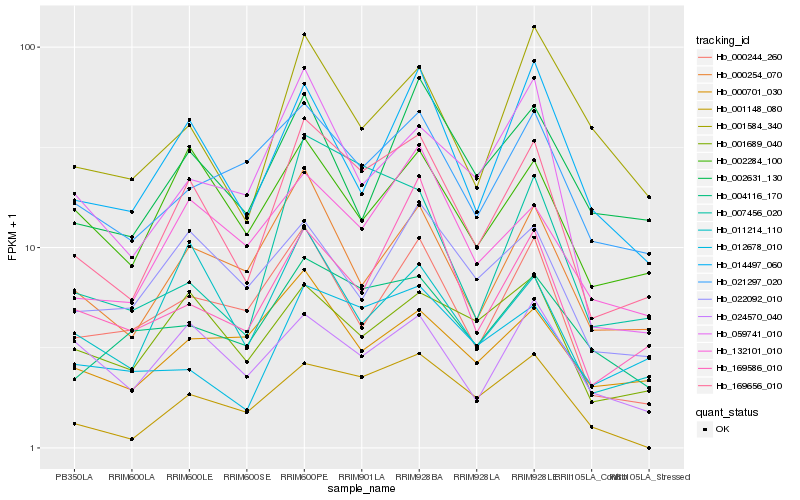
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169656_010 |
0.0 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_000254_070 |
0.1359168503 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 3 |
Hb_132101_010 |
0.143069394 |
- |
- |
hypothetical protein B456_007G078100 [Gossypium raimondii] |
| 4 |
Hb_011214_110 |
0.1479675123 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 5 |
Hb_014497_060 |
0.1500177641 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 6 |
Hb_001689_040 |
0.1503146844 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_000244_260 |
0.1512703849 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002284_100 |
0.1514204388 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 9 |
Hb_004116_170 |
0.1532864949 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_021297_020 |
0.1545587305 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 11 |
Hb_007456_020 |
0.1545829791 |
- |
- |
unknown [Lotus japonicus] |
| 12 |
Hb_001148_080 |
0.1594307698 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 13 |
Hb_059741_010 |
0.1617048159 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_012678_010 |
0.1621848855 |
- |
- |
PREDICTED: outer envelope protein 61-like [Gossypium raimondii] |
| 15 |
Hb_000701_030 |
0.1630583631 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 16 |
Hb_022092_010 |
0.1633942895 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 17 |
Hb_002631_130 |
0.165047967 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 18 |
Hb_001584_340 |
0.1659967816 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 19 |
Hb_024570_040 |
0.1661028195 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 20 |
Hb_169586_010 |
0.1681902692 |
- |
- |
Acid phosphatase/vanadium-dependent haloperoxidase-related protein [Theobroma cacao] |