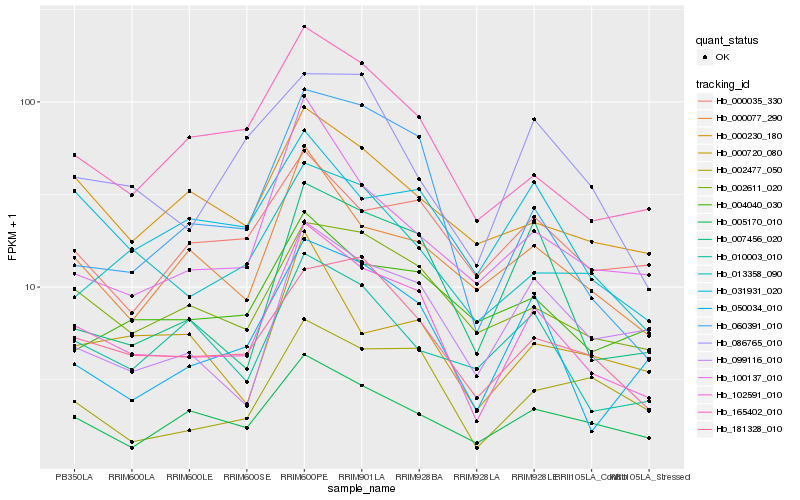
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102591_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 2 |
Hb_165402_010 |
0.1373071173 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 3 |
Hb_181328_010 |
0.1399636819 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_050034_010 |
0.1407873324 |
- |
- |
PREDICTED: cullin-1 [Jatropha curcas] |
| 5 |
Hb_002611_020 |
0.1408627936 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 6 |
Hb_000077_290 |
0.143145491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007456_020 |
0.1467607014 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_005170_010 |
0.1477233332 |
- |
- |
- |
| 9 |
Hb_013358_090 |
0.1478112827 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 10 |
Hb_031931_020 |
0.1500026531 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 11 |
Hb_060391_010 |
0.1511146829 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_099116_010 |
0.1515289538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000230_180 |
0.1586910319 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 14 |
Hb_100137_010 |
0.1610880335 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 15 |
Hb_010003_010 |
0.1642800413 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004040_030 |
0.1643828262 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002477_050 |
0.1651738641 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_000035_330 |
0.1689431431 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 19 |
Hb_086765_010 |
0.1701297218 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 20 |
Hb_000720_080 |
0.1719770108 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |