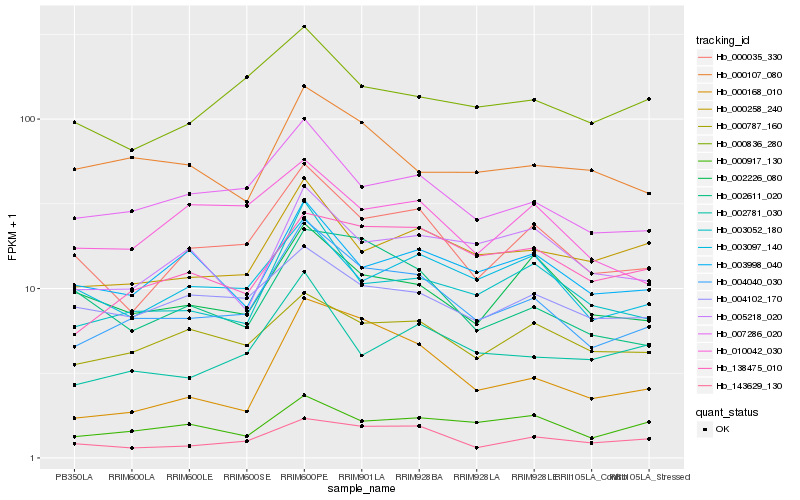
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004040_030 |
0.0 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 2 |
Hb_007286_020 |
0.0935171832 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 3 |
Hb_003052_180 |
0.1066333467 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 4 |
Hb_000035_330 |
0.1071731652 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 5 |
Hb_143629_130 |
0.1221405728 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_002781_030 |
0.1235908531 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000258_240 |
0.1236371075 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 8 |
Hb_000168_010 |
0.1279791229 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RFWD3-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000787_160 |
0.1304006613 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_138475_010 |
0.1304271307 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Populus euphratica] |
| 11 |
Hb_003097_140 |
0.1308875306 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 12 |
Hb_002611_020 |
0.1319105975 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 13 |
Hb_000917_130 |
0.1319709384 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 14 |
Hb_002226_080 |
0.1323088181 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000107_080 |
0.133139967 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 16 |
Hb_003998_040 |
0.1333398582 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_000836_280 |
0.1339892267 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010042_030 |
0.134333464 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 19 |
Hb_004102_170 |
0.1343747954 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 20 |
Hb_005218_020 |
0.1347369642 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |