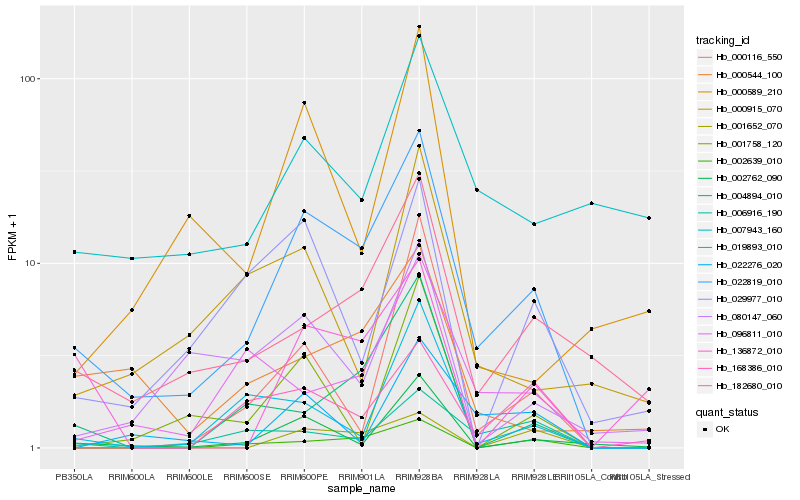
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022819_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_182680_010 |
0.235298652 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 3 |
Hb_002762_090 |
0.2465231233 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_002639_010 |
0.2494961606 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 5 |
Hb_000544_100 |
0.2647956387 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 6 |
Hb_004894_010 |
0.2674068781 |
- |
- |
- |
| 7 |
Hb_096811_010 |
0.2710344543 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 8 |
Hb_006916_190 |
0.2719220597 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_019893_010 |
0.2768328785 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 10 |
Hb_080147_060 |
0.2775973094 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029977_010 |
0.2794347029 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_007943_160 |
0.2807236323 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 13 |
Hb_022276_020 |
0.2828568077 |
- |
- |
- |
| 14 |
Hb_000589_210 |
0.2832012423 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 15 |
Hb_001652_070 |
0.2869588804 |
- |
- |
PREDICTED: uncharacterized protein LOC103439285 [Malus domestica] |
| 16 |
Hb_000116_550 |
0.2877934258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_168386_010 |
0.2892197985 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_001758_120 |
0.2906566365 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_136872_010 |
0.2908586732 |
- |
- |
serine/threonine-specific receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_000915_070 |
0.2920063934 |
- |
- |
ATP binding protein, putative [Ricinus communis] |