| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
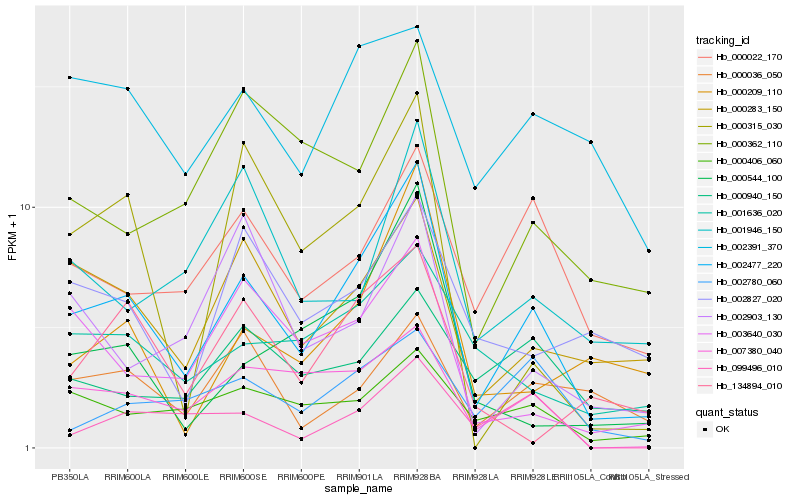
Hb_002477_220 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 2 |
Hb_000283_150 |
0.1944459951 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 3 |
Hb_007380_040 |
0.1974586526 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_000406_060 |
0.20978901 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 5 |
Hb_000022_170 |
0.2127658806 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 6 |
Hb_003640_030 |
0.213474032 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001636_020 |
0.214291175 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 8 |
Hb_000209_110 |
0.2161775373 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 9 |
Hb_000362_110 |
0.2182248357 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 10 |
Hb_000544_100 |
0.2197850825 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 11 |
Hb_000315_030 |
0.2264101547 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 12 |
Hb_002780_060 |
0.2276256564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001946_150 |
0.2323017635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000036_050 |
0.2359704863 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 15 |
Hb_000940_150 |
0.23606506 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 16 |
Hb_002391_370 |
0.236401042 |
- |
- |
PREDICTED: aconitate hydratase 1 [Jatropha curcas] |
| 17 |
Hb_002903_130 |
0.2364510298 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 18 |
Hb_099496_010 |
0.2369010703 |
- |
- |
PREDICTED: uncharacterized protein LOC105138000 [Populus euphratica] |
| 19 |
Hb_134894_010 |
0.2372173241 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 20 |
Hb_002827_020 |
0.2385123259 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |