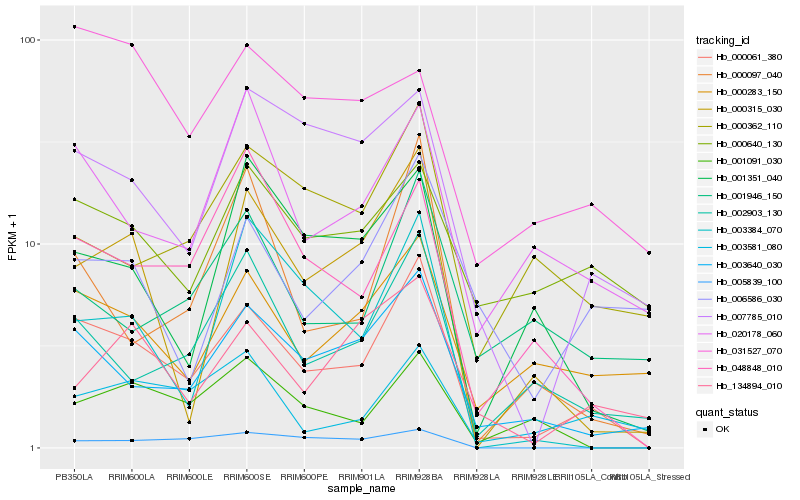
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_380 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003640_030 |
0.1697348828 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002903_130 |
0.1946597848 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 4 |
Hb_000283_150 |
0.2032211679 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 5 |
Hb_048848_010 |
0.2055402957 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 6 |
Hb_000315_030 |
0.2087412606 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 7 |
Hb_020178_060 |
0.2135181287 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 8 |
Hb_000097_040 |
0.2172102921 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 9 |
Hb_007785_010 |
0.2209811545 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |
| 10 |
Hb_003581_080 |
0.2212534131 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 11 |
Hb_001091_030 |
0.2238896188 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 12 |
Hb_005839_100 |
0.229890368 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 13 |
Hb_003384_070 |
0.2338266589 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 14 |
Hb_001351_040 |
0.2348522457 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000362_110 |
0.2370948797 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_001946_150 |
0.237506459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000640_130 |
0.2378588168 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_134894_010 |
0.2391268872 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 19 |
Hb_006586_030 |
0.2453040037 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 20 |
Hb_031527_070 |
0.2520172454 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |