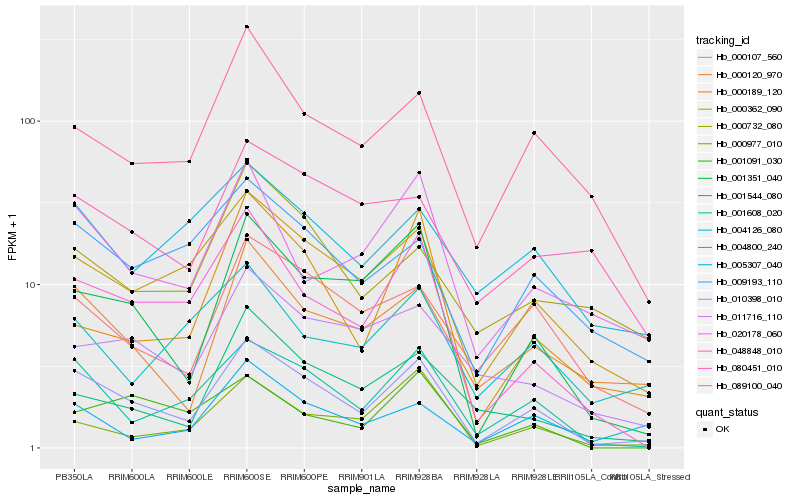
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010398_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 2 |
Hb_001608_020 |
0.1259636989 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000362_090 |
0.145241649 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001351_040 |
0.149495026 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_048848_010 |
0.1500749961 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 6 |
Hb_000120_970 |
0.1594614531 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000107_560 |
0.1718753064 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 8 |
Hb_089100_040 |
0.1740267559 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 9 |
Hb_009193_110 |
0.1741455855 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_020178_060 |
0.1755347747 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 11 |
Hb_000977_010 |
0.1904826353 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 12 |
Hb_011716_110 |
0.1935604148 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 13 |
Hb_004800_240 |
0.1941975303 |
- |
- |
PREDICTED: vam6/Vps39-like protein [Jatropha curcas] |
| 14 |
Hb_005307_040 |
0.1987017595 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 15 |
Hb_001544_080 |
0.2007402047 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 16 |
Hb_001091_030 |
0.2009444694 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 17 |
Hb_000189_120 |
0.2013814663 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 18 |
Hb_004126_080 |
0.202498291 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 19 |
Hb_080451_010 |
0.2041274838 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 20 |
Hb_000732_080 |
0.2045571954 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |