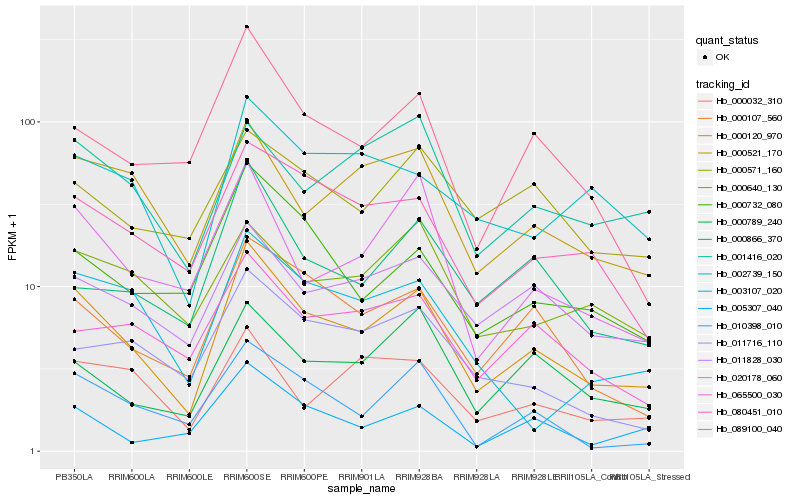
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_970 |
0.0 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000107_560 |
0.1374633727 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 3 |
Hb_080451_010 |
0.1481829875 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680 [Jatropha curcas] |
| 4 |
Hb_000521_170 |
0.1486609185 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 5 |
Hb_011828_030 |
0.1498165736 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 6 |
Hb_020178_060 |
0.1560541257 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 7 |
Hb_089100_040 |
0.1590056962 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 8 |
Hb_010398_010 |
0.1594614531 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 9 |
Hb_000789_240 |
0.1595579935 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 10 |
Hb_000732_080 |
0.1621641135 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g45440 [Jatropha curcas] |
| 11 |
Hb_065500_030 |
0.1661212352 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 [Jatropha curcas] |
| 12 |
Hb_002739_150 |
0.1684786801 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 13 |
Hb_000866_370 |
0.1684977408 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 14 |
Hb_005307_040 |
0.1686237974 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 15 |
Hb_000032_310 |
0.1706532112 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_001416_020 |
0.1714920559 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 17 |
Hb_000640_130 |
0.1715943547 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003107_020 |
0.1729404884 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 19 |
Hb_011716_110 |
0.1730473425 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 20 |
Hb_000571_160 |
0.1749661572 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |