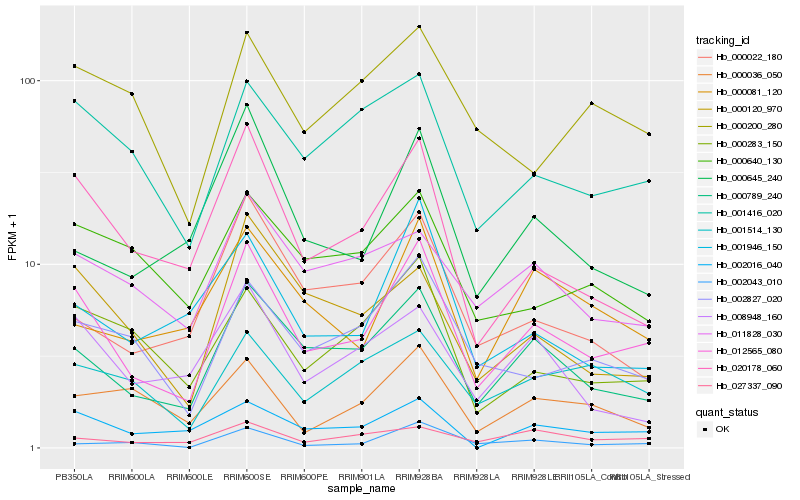
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012565_080 |
0.0 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_000789_240 |
0.1254792233 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 3 |
Hb_020178_060 |
0.1484564562 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 4 |
Hb_001416_020 |
0.1628611785 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 5 |
Hb_002827_020 |
0.1659217584 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 6 |
Hb_000283_150 |
0.1702129762 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 7 |
Hb_002016_040 |
0.1711913562 |
rubber biosynthesis |
Gene Name: SRPP8 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 8 |
Hb_001946_150 |
0.1764606479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000120_970 |
0.1802614095 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000645_240 |
0.1812482777 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 11 |
Hb_002043_010 |
0.1813631727 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_008948_160 |
0.1817357339 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 13 |
Hb_001514_130 |
0.1818410445 |
- |
- |
hypothetical protein JCGZ_16073 [Jatropha curcas] |
| 14 |
Hb_000081_120 |
0.1825081854 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 15 |
Hb_027337_090 |
0.1849192666 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 16 |
Hb_000022_180 |
0.1862971807 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000640_130 |
0.1863027061 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_011828_030 |
0.1867580229 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 19 |
Hb_000036_050 |
0.1888523797 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6-like [Camelina sativa] |
| 20 |
Hb_000200_280 |
0.1915983772 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |