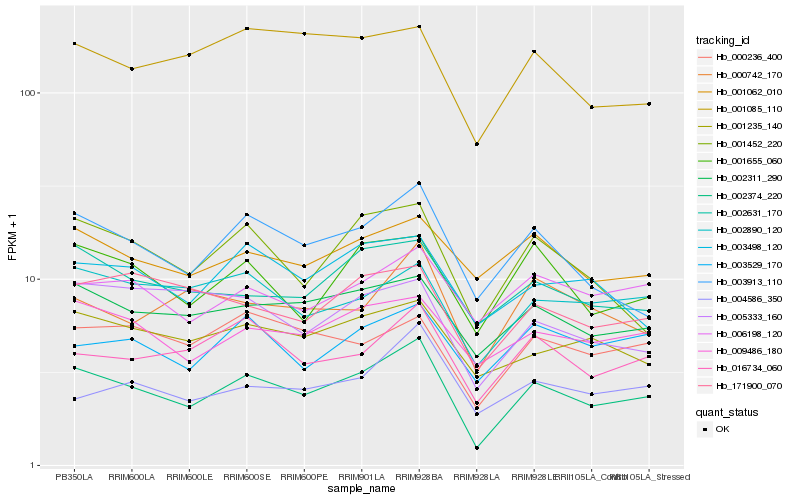
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_220 |
0.0 |
- |
- |
hypothetical protein CICLE_v10001597mg [Citrus clementina] |
| 2 |
Hb_003913_110 |
0.1121417207 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 3 |
Hb_002311_290 |
0.1161020512 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 4 |
Hb_001655_060 |
0.1180715922 |
- |
- |
PREDICTED: RNA-binding protein 28 [Jatropha curcas] |
| 5 |
Hb_016734_060 |
0.1186399472 |
- |
- |
PREDICTED: nucleolar protein 6 [Jatropha curcas] |
| 6 |
Hb_009486_180 |
0.1210880611 |
- |
- |
unknown [Glycine max] |
| 7 |
Hb_001452_220 |
0.1222845288 |
- |
- |
PREDICTED: molybdopterin biosynthesis protein CNX1 [Jatropha curcas] |
| 8 |
Hb_002890_120 |
0.1248119431 |
- |
- |
Nucleic acid binding,RNA binding isoform 1 [Theobroma cacao] |
| 9 |
Hb_002631_170 |
0.125302757 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 10 |
Hb_000236_400 |
0.1270392055 |
- |
- |
hypothetical protein JCGZ_01058 [Jatropha curcas] |
| 11 |
Hb_001235_140 |
0.1281866705 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 12 |
Hb_171900_070 |
0.1304864348 |
- |
- |
- |
| 13 |
Hb_003498_120 |
0.1314589015 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 14 |
Hb_003529_170 |
0.131580541 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX29 [Jatropha curcas] |
| 15 |
Hb_004586_350 |
0.1332533988 |
- |
- |
Phosphoribosylaminoimidazole-succinocarboxamide synthase [Morus notabilis] |
| 16 |
Hb_000742_170 |
0.133579017 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_006198_120 |
0.1345788553 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 18 |
Hb_001085_110 |
0.135809187 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 19 |
Hb_001062_010 |
0.1359087632 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 20 |
Hb_005333_160 |
0.137444498 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |