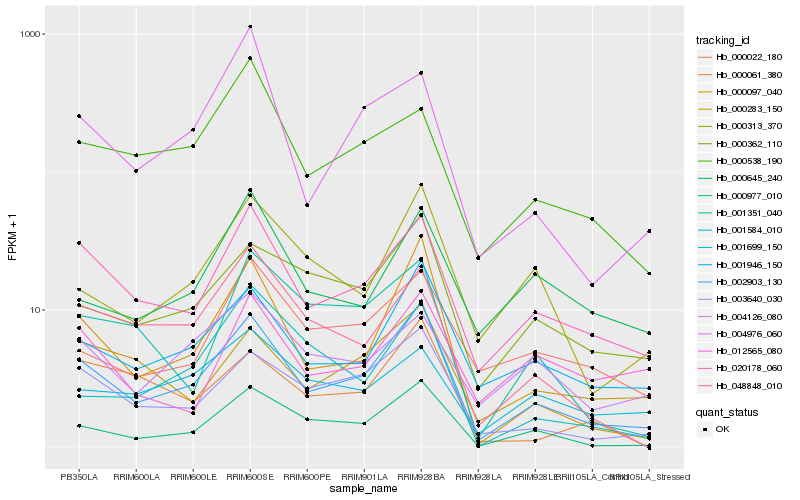
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002903_130 |
0.0 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 2 |
Hb_000977_010 |
0.1152425786 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 3 |
Hb_020178_060 |
0.1212297571 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 4 |
Hb_001946_150 |
0.1312102429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003640_030 |
0.1329604705 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000097_040 |
0.1384496075 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 7 |
Hb_000362_110 |
0.1524044473 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_000313_370 |
0.1627072394 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_048848_010 |
0.1731757406 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 10 |
Hb_000022_180 |
0.1817459708 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000283_150 |
0.1841593647 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 12 |
Hb_001351_040 |
0.1881790711 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004126_080 |
0.1915446405 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 14 |
Hb_000645_240 |
0.1930740528 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 15 |
Hb_000061_380 |
0.1946597848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004976_060 |
0.1956667783 |
- |
- |
- |
| 17 |
Hb_000538_190 |
0.1975185682 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 18 |
Hb_001699_150 |
0.1983375599 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 19 |
Hb_012565_080 |
0.2024348474 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 20 |
Hb_001584_010 |
0.2044535635 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |