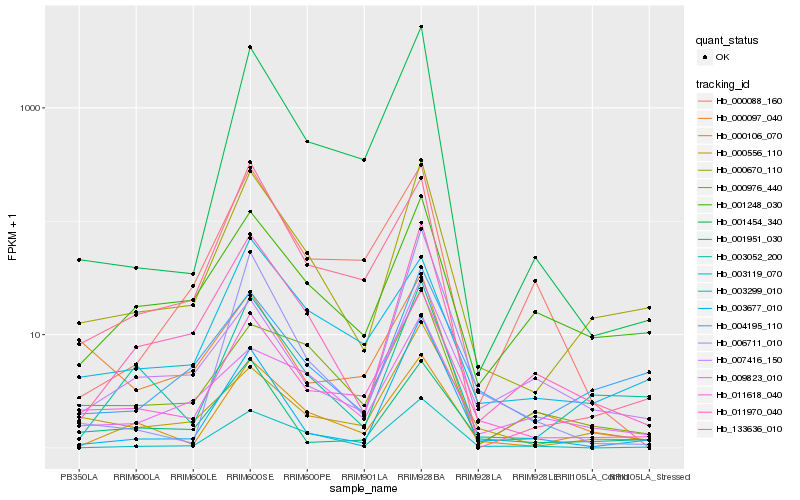
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000556_110 |
0.1328400121 |
- |
- |
hypothetical protein POPTR_0014s06790g [Populus trichocarpa] |
| 3 |
Hb_000670_110 |
0.1476150897 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 4 |
Hb_000106_070 |
0.1604633302 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 5 |
Hb_001454_340 |
0.1634251287 |
- |
- |
ATDI21 [Manihot esculenta] |
| 6 |
Hb_003119_070 |
0.1776486631 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 7 |
Hb_007416_150 |
0.1786899399 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 8 |
Hb_011970_040 |
0.1796680871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001951_030 |
0.1798980817 |
- |
- |
hypothetical protein JCGZ_04584 [Jatropha curcas] |
| 10 |
Hb_000976_440 |
0.1856673108 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001248_030 |
0.1872977332 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000097_040 |
0.1879997526 |
transcription factor |
TF Family: MYB |
myb domain protein [Hevea brasiliensis] |
| 13 |
Hb_003677_010 |
0.1896296043 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 14 |
Hb_009823_010 |
0.1897072641 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 15 |
Hb_004195_110 |
0.1902294503 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 16 |
Hb_006711_010 |
0.1922647195 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 17 |
Hb_133636_010 |
0.1950990312 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 18 |
Hb_003299_010 |
0.1992569516 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 19 |
Hb_003052_200 |
0.2027551107 |
- |
- |
hypothetical protein POPTR_0010s14360g [Populus trichocarpa] |
| 20 |
Hb_000088_160 |
0.2040302544 |
- |
- |
PREDICTED: uncharacterized protein LOC105629775 [Jatropha curcas] |